Scientific Computing and Data / Mount Sinai Data Warehouse / Somatic Genomic Test Results
Somatic Genomic Test Results
Background
- Scientific Computing and Data, Mount Sinai Innovation Partners, the Department of Molecular Pathology, and Tisch Cancer Institute have partnered with genomic testing vendors to develop a genomic research data warehouse comprised of somatic genomic testing results ordered at all Mount Sinai facilities.
- All somatic genomic tests performed by Foundation Medicine and Caris for Mount Sinai patients are hosted on MSDW in multiple data formats (see table below), encompassing both structured and raw genomic data.
| Vendor | Patient Demographics | File Format |
# of Files (as of 11/20/2024) |
| Foundation Medicine |
Female: 54% Age (mean, years): 61 |
8,368 | |
| xml | 8,368 | ||
| json | 3,090 | ||
| vcf | 7,351 | ||
| bam | 12,479 | ||
| Caris |
Female: 45% Age (mean, years): 64 |
1,045 | |
| xml | 1,045 | ||
| json | 1,044 | ||
| vcf | 814 | ||
| bam | 1,624 | ||
| fastq | 1,241 |
- Somatic genomic testing is used in oncology to identify genetic mutations in cancer cells.
- The somatic genomic testing results in this data set represent a wide variety of solid tumors and hematologic malignancies from a range of organ systems and tissues including breast, lung, bladder, ovary, and liver.
How the Somatic Genomic Results are Returned to Mount Sinai
- Data flows from the external lab to Minerva, Mount Sinai’s high-performance computer (see illustration below).
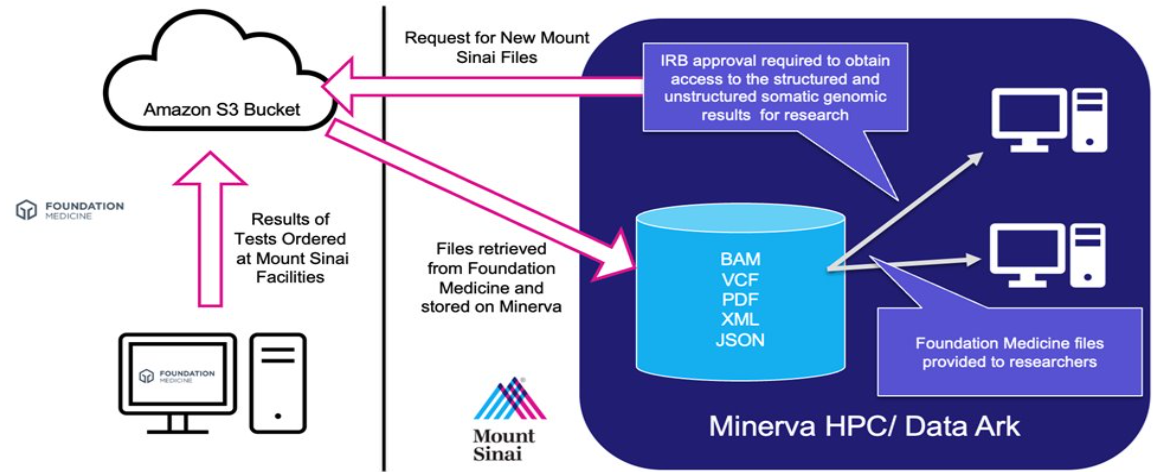
- Minerva retrieves new results from a secure Amazon S3 bucket multiple times daily.
Access
- The somatic genomic results include PHI (Protected Health Information), so access to this data requires an approved Institutional Review Board (IRB) protocol outlining the somatic genomic results to be studied.
- Data requests go through Mount Sinai Data Warehouse at MSDW support.
- The MSDW team will work with the Mount Sinai researcher to identify the required results, as outlined in the IRB protocol.
- If the study requires analysis of the raw genomic data, the MSDW team will be notified and will provision the access to the necessary BAM and FASTQ files hosted under MSDW to research staff listed on the IRB protocol.
MSDW Data Sets
- Please visit the MSDW webpage to explore other data sets.
