Scientific Computing and Data / Mount Sinai Data Warehouse / About MSDW
About The Mount Sinai Data Warehouse
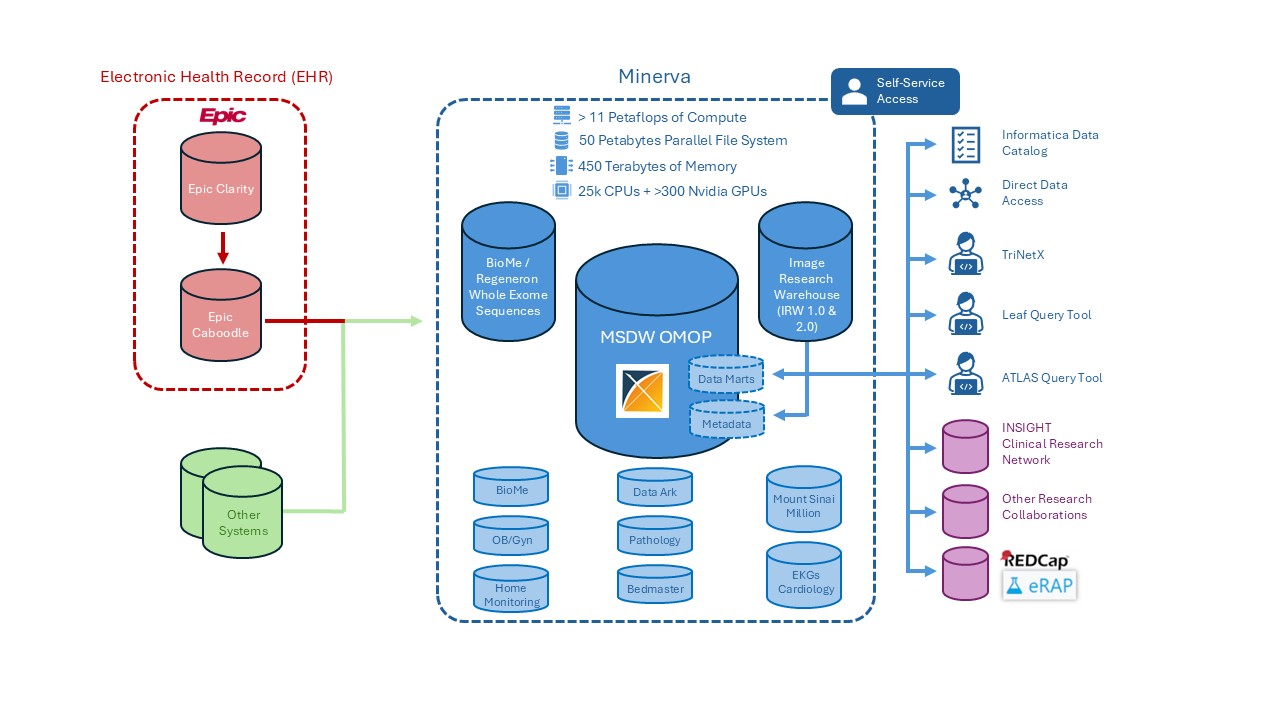
The Mount Sinai Data Warehouse (MSDW) collects clinical and operational data for use in clinical and translational research, as well as quality and improvement initiatives.
Mount Sinai Data Warehouse Ecosystem
Click here to read more about data sources, transactions, and comprehensive descriptions of data in the Data Warehouse. MSDW is approved by the IRB.
For more resources, presentations, training materials, and documentation, click here.
MSDW Data
The MSDW provides researchers access to data on patients in the Mount Sinai Health System:
- Over 12 million patient records
- Over 105 million patient encounters
The majority of the data collected by the MSDW comes from the Epic Clarity and Caboodle databases, as Epic is the primary electronic health record (EHR) across the Mount Sinai Health System (MSHS).
Click here for more details on MSDW’s OMOP Data Contents.
Click here to find out about the somatic genomic test results data.
AIR·MS (Artificial Intelligence-Ready Mount Sinai)
Both identifiable and de-identified versions of MSDW datasets are offered in AIR·MS, a cloud-based, multi-modal health data platform that integrates patient data generated from different clinical departments across Health Systems. Click here to find out more about AIR·MS
Mount Sinai Facility |
Epic Go-Live |
| Mount Sinai Hospital (MSH) | 2011 |
| Mount Sinai Queens | 2013 |
| Mount Sinai West | 2018 |
| Mount Sinai Morningside | 2018 |
| Mount Sinai Brooklyn | 2019 |
| Mount Sinai Beth Israel | 2020 |
The clinical data are extracted from Mount Sinai’s Epic Caboodle database and other ancillary systems, transformed into the OMOP Common Data Model (CDM) format, and loaded to the MSDW database. These data are updated daily.
The MSDW is situated on the Minerva High-Performance Computing (HPC) cluster alongside other research data sets.
Supported by grant UL1TR004419 from the National Center for Advancing Translational Sciences, National Institutes of Health.
