Scientific Computing and Data / High Performance Computing / Documentation / R and Rstudio
R and Rstudio
Contents
R
Rstudio
Rstudio Connect Server
R
R is a widely used open-source software for statistical computing and graphics supported by the R Core Team and the R Foundation for Statistical Computing. Many versions of R are available to use on Minerva. To see a list of installed versions simply type:
$ module -r spider “^R$”where “^R$” is a regular expression meaning that the module name starts with R and ends with R as well.
Note that new versions of R are periodically added and the default version of R changes accordingly. You may want to explicitly specify the version of R in your module load command (i.e. module load R/3.5.3) to avoid picking up a new version of R when you don’t want it. We would recommend users to use the most recent version installed. Our current R installation comes with a large number of popular scientific and high-performance computing packages preinstalled, including Bioconductor packages.
Usage
R may be run either interactively via the R terminal, or as a batch process reading commands from a script file. To launch the R terminal, open an interactive session by submitting a job to the interactive LSF queue. Then simply execute the R command. After the terminal is launched in the interactive mode, users can run R commands at the prompt:
[choh07@li04e01 ~]$ bsub -q interactive -P acc_hpcstaff -n 1 -W 1:00 -R rusage[mem=8000] -XF -Is /bin/bash Job <65508400> is submitted to queue . <> <> <> [choh07@lc03e16 ~]$ ml R/4.2.0 [choh07@lc03e16 ~]$ R R version 4.2.0 (2022-04-22) -- "Vigorous Calisthenics" Copyright (C) 2022 The R Foundation for Statistical Computing Platform: x86_64-pc-linux-gnu (64-bit) R is free software and comes with ABSOLUTELY NO WARRANTY. You are welcome to redistribute it under certain conditions. Type 'license()' or 'licence()' for distribution details. Natural language support but running in an English locale R is a collaborative project with many contributors. Type 'contributors()' for more information and 'citation()' on how to cite R or R packages in publications. Type 'demo()' for some demos, 'help()' for on-line help, or 'help.start()' for an HTML browser interface to help. Type 'q()' to quit R. > print("Hello World!", quote = FALSE) [1] Hello World! >
Rscript, on the other hand, can be used to run a R script in which a sequence of R commands has been saved. The syntax is:
$ Rscript <options> <your R script>
For example,
$ cat hello.R print("Hello World!", quote = FALSE) $ Rscript hello.R [1] Hello World!
Installing New Packages Locally
If you find that a particular package you need is missing from the R version you use, you may install it yourself into your local space. In some cases you will need to build/maintain your own set of R packages in your space locally due to various reasons.
To list currently installed packages, typing library() from the R command prompt, as shown.
$ R -e 'library()'
There are multiple ways to install R packages. Here we present one of them below as an example. To begin, you need to create a directory in your local space to install R packages into. We will use “~/.Rlib” in this example, which is the default location and its path is predefined in the environment variable “R_LIBS_USER” but you may choose as you wish.
$ mkdir ~/.Rlib
When using a different R version, it is recommended to reinstall your existing packages to ensure compatibility and optimal performance with the updated R environment. This process helps prevent potential conflicts and ensures that all packages are compiled specifically for the new R version’s architecture and dependencies.
To make R packages installed into this local directory, create the “.Renviron” file in your home directory to redefine the environment variable “R_LIBS” for the R packages search path accordingly:
$ cat ~/.Renviron R_LIBS=${R_LIBS_USER}:${R_LIBS}
Note that the path to your local R packages directory is prepended to the system one in order to point R to the directory where your local packages are installed first. Load R and start up an R session from the terminal. You should be able to use the standard R command to install the package interactively now.
$ R R version 4.2.0 (2022-04-22) -- "Vigorous Calisthenics" Copyright (C) 2022 The R Foundation for Statistical Computing Platform: x86_64-pc-linux-gnu (64-bit) . . . > .libPaths() [1] "/hpc/users/choh07/.Rlib" [2] "/hpc/packages/minerva-centos7/rpackages/4.2.0/site-library" [3] "/hpc/packages/minerva-centos7/rpackages/bioconductor/3.15" [4] "/hpc/packages/minerva-centos7/R/4.2.0/lib64/R/library" > install.packages("zoo") Installing package into ‘/hpc/users/choh07/.Rlib’ (as ‘lib’ is unspecified) . . . ** R ** demo ** inst ** byte-compile and prepare package for lazy loading ** help *** installing help indices ** building package indices ** installing vignettes ** testing if installed package can be loaded from temporary location ** checking absolute paths in shared objects and dynamic libraries ** testing if installed package can be loaded from final location ** testing if installed package keeps a record of temporary installation path * DONE (zoo) >
Rstudio
RStudio is an integrated development environment (IDE) for R. It includes a console, syntax-highlighting editor that supports direct code execution, as well as tools for plotting, history, debugging and workspace management. It is available in two formats: RStudio Desktop is a regular desktop application while RStudio Server runs on a remote server and allows accessing RStudio using a web browser. RStudio Desktop and RStudio Server are both available in free and fee-based (commercial) editions.
On Minerva, the recommended ways of accessing Rstudio are running Rstudio Server via Open OnDemand or one simple command to get On-the-fly Rstudio over Web in a Minerva job.
Using Rstudio Server via Open OnDemand
To access Rstudio Server via OnDemand, please
- Authenticate into Open OnDemand on Minerva
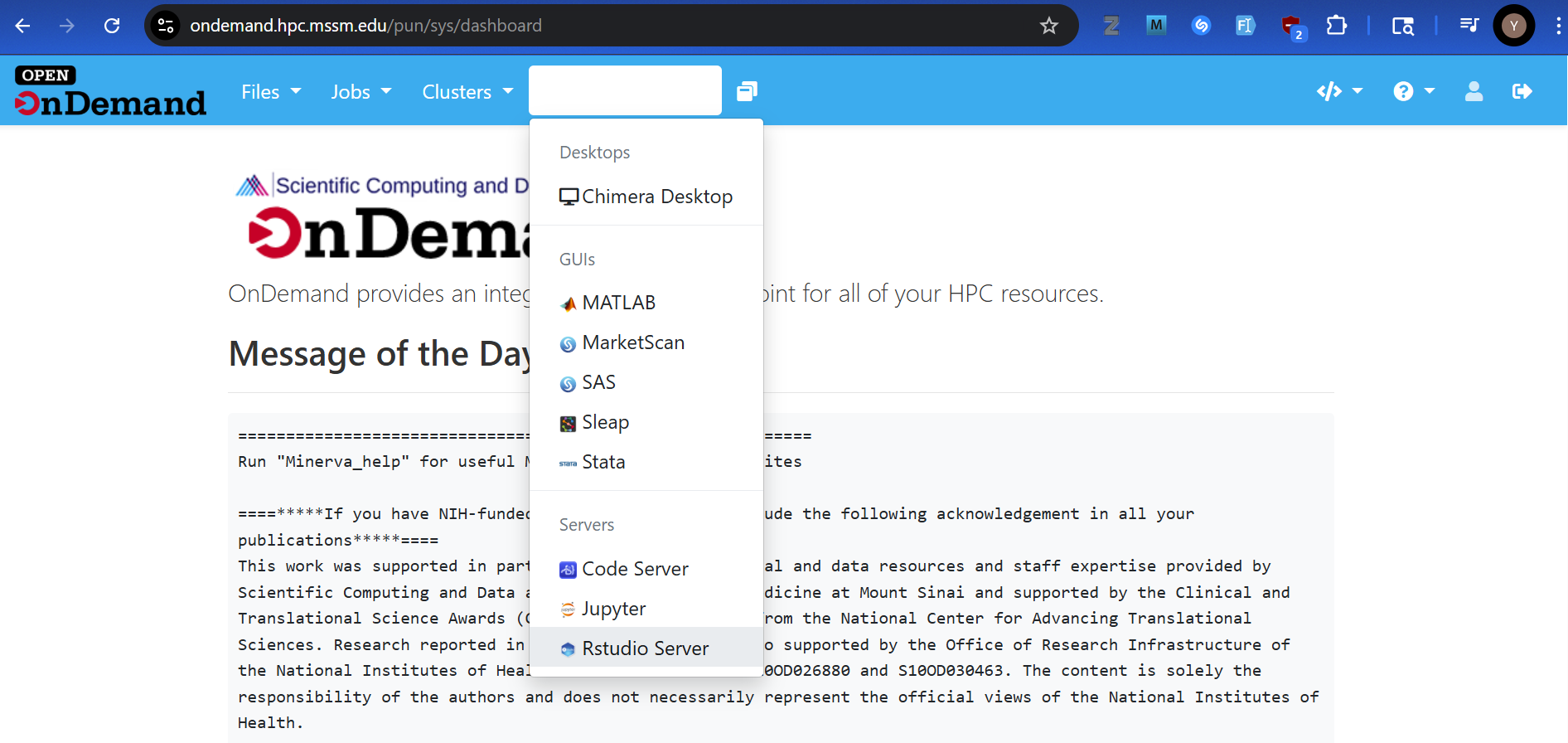
- Navigate to RStudio Server app
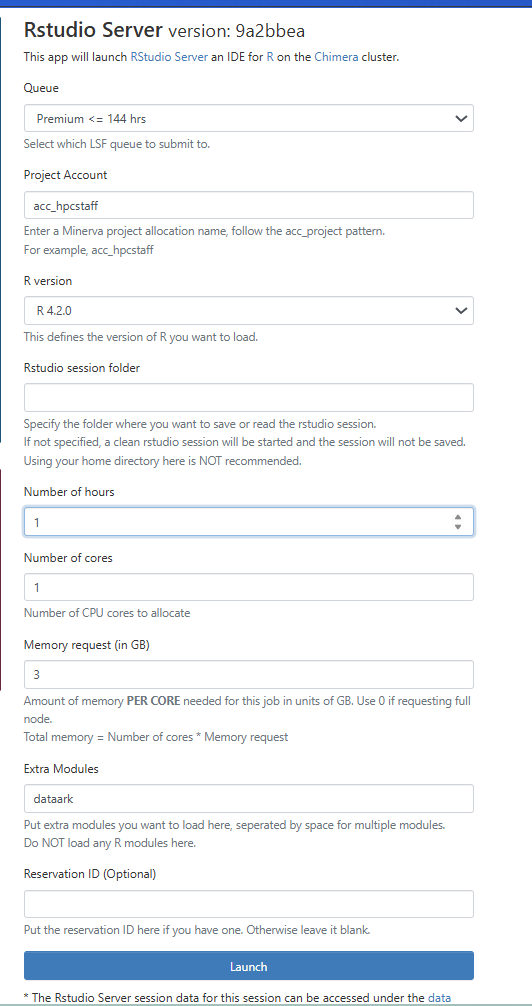
- Complete the job resource request for the LSF job
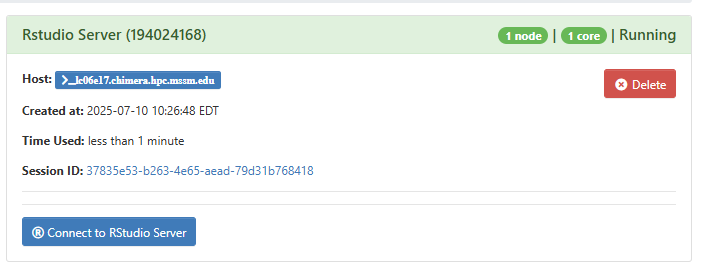
- Launch the application
Using Rstudio Server via the On-the-fly Script
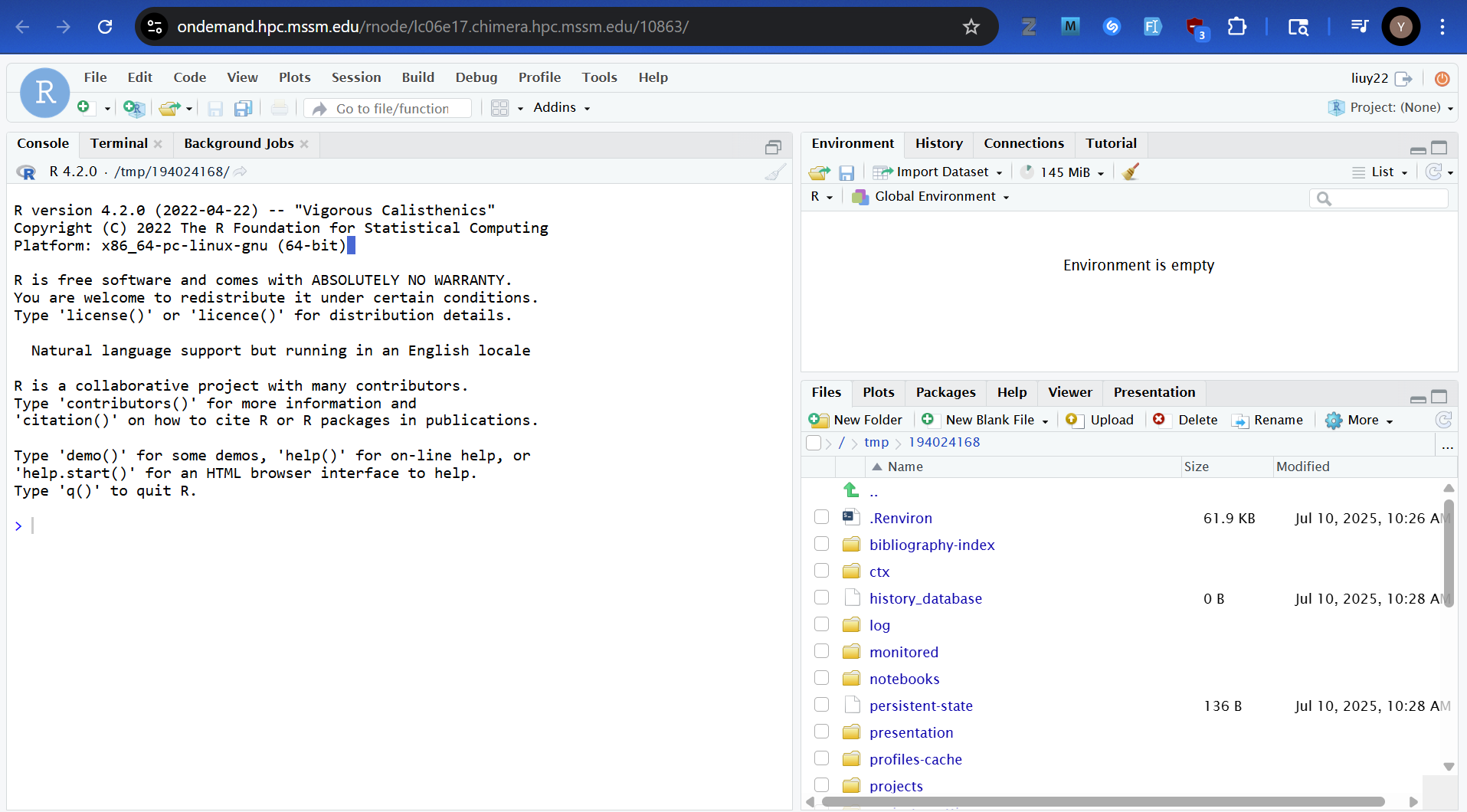
We have developed simple tool scripts for users to launch RStudio server with interactive web interface inside the LSF job with dedicated resources. With one simple command,you can run the free Rstudio Server remotely on Minerva and access the RStudio interface using your local web browser.
The script is located in the login nodes under
/usr/local/bin/minerva-rstudio-web-module.shWith one command on the terminal, you will launch RStudio server and access interactive web interface inside the LSF job with dedicated resources on Minerva.
Usage:
For example, to start an RStudio web session with R, on the login nodes, run commands minerva-rstudio-web-module.sh (using R 4.2.0) with default resource configuration and URL to access it. Please see the -h option for help messages containing resource requests and installing packages.
Example:
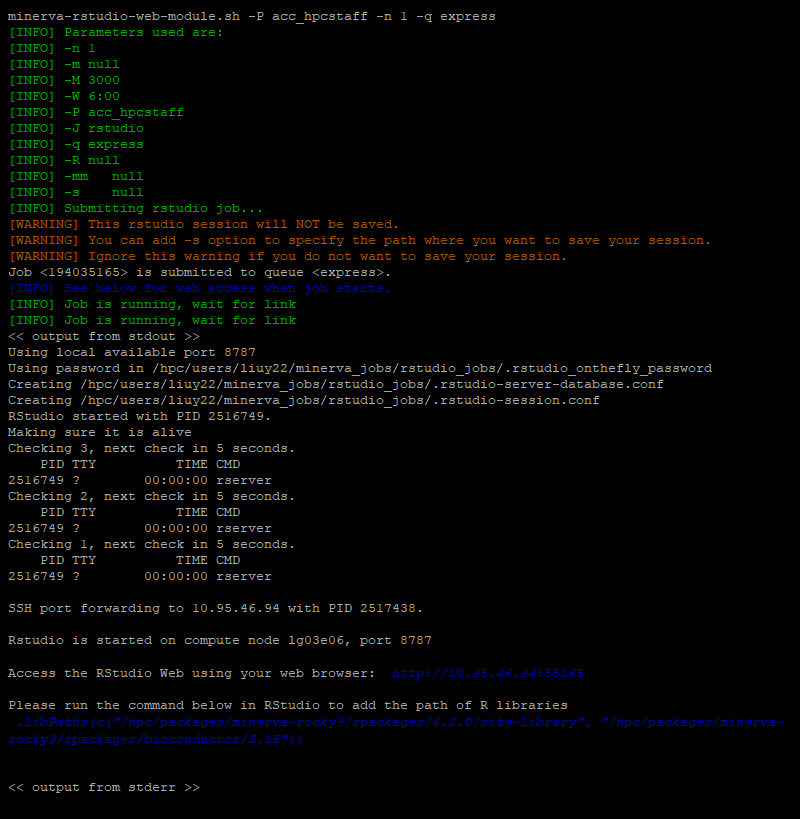
The RStudio web interface will be available at URL http://10.95.46.94:55165. Use the browser on your laptop to access it. Here 10.95.46.94: is the login node IP on the campus network and 55165 is the port. 5165 is the last 4 digit of the job 194035165 running the instance.
What happens behind the scene? This tool wraps the following tasks in one command.
- writes and submits an LSF job script to launch the RStudio Server
- provides the URL link to access the instance
See the Package Installations section in the help message for packages not installed.
Installing R Modules That Require External Libraries From Minerva Packages for on-the-fly RStudio Web
Occasionally, you may need to install an R module that requires one or more dynamic libraries from Minerva packages. In these cases, you should install the R module into the private library that is used by rstudio outside of rstudio using hard-coded paths to those libraries.
A convenient way to signal the linker that it should encode the full path to dynamic libraries is to set LD_RUN_PATH to LD_LIBRARY_PATH after you load all the Minerva packages needed to build the R module but before you actually build the module. When you build the module, you will need to specify the destination library as the default private library for rstudio.
Example:
ml R/4.2.0
ml geos
export LD_RUN_PATH=$LD_LIBRARY_PATH
R
> install.packages(“Seurat”, lib=”~/.Rlib”)
RStudio/Posit Connect Server
What is Rstudio/Posit Connect?
Rstudio/Posit Connect server is available on Minerva since Aug 2020, where you can publish Shiny, R Markdown and Jupyter for collaborators or others. See more details about Rstudio Connect here.
How many users allowed to log into Rstudio Connect?
Currently, the subscription is upgraded to allow 45 named users on the server.
How to publish?
Rstudio-connect server is at https://rstudio-connect.hpc.mssm.edu
To publish your R product with Rstudio-connect:
1. Login to Rstudio-connect server using your Sinai username and password associated with your school account followed immedately by the 6-digit VIP token.
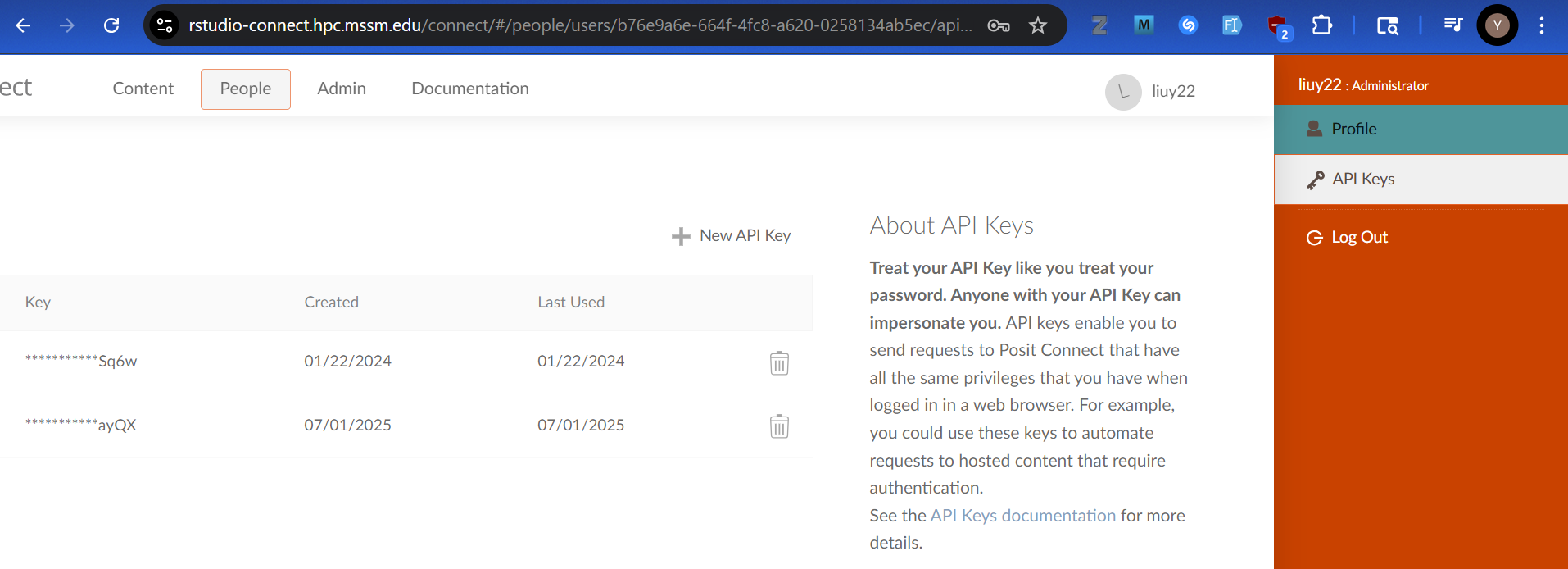
2. Generate an API key, if not already, and save the API key value elsewhere for later use.
3. Login to Minerva by ssh -X userid@minerva.hpc.mssm.edu ( X11 forwarding needed for GUI).
4. After login, navigate to the application directory and ensure this directory includes all necessary files (e.g., app.R, ui.R, server.R, or other dependencies).
5. Ensure the “rsconnect” R package has already been installed.
6. Register the Rstuido Connect server with “rsconnect”, if not already.
$ ml R/4.2.0 $ R -q > library(rsconnect) > rsconnect::addConnectServer("https://rstudio-connect.hpc.mssm.edu", "myServer") > rsconnect::connectApiUser( server = "myServer", account = "<Sinai_username>", apiKey = "<Connect_API_key>" # described in step #2 )
7. Leveraging existing functions (such as deployApp) of the “rsconnect” package to deply your Shiny app, R Markdown and various types of APIs to the Rstudio Connect server.
> rsconnect::deployApp( appDir = "path/to/your/app", appName = "your_app_name", appTitle = "Your App Title", server = "myServer" )
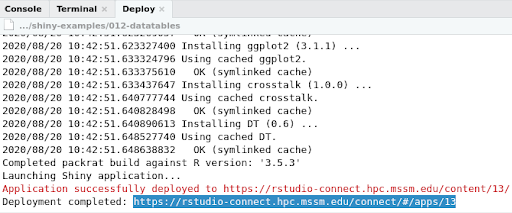
8. After your application is successfully deployed as shown in the deploy log, you can copy the URL and open it in any browser you like
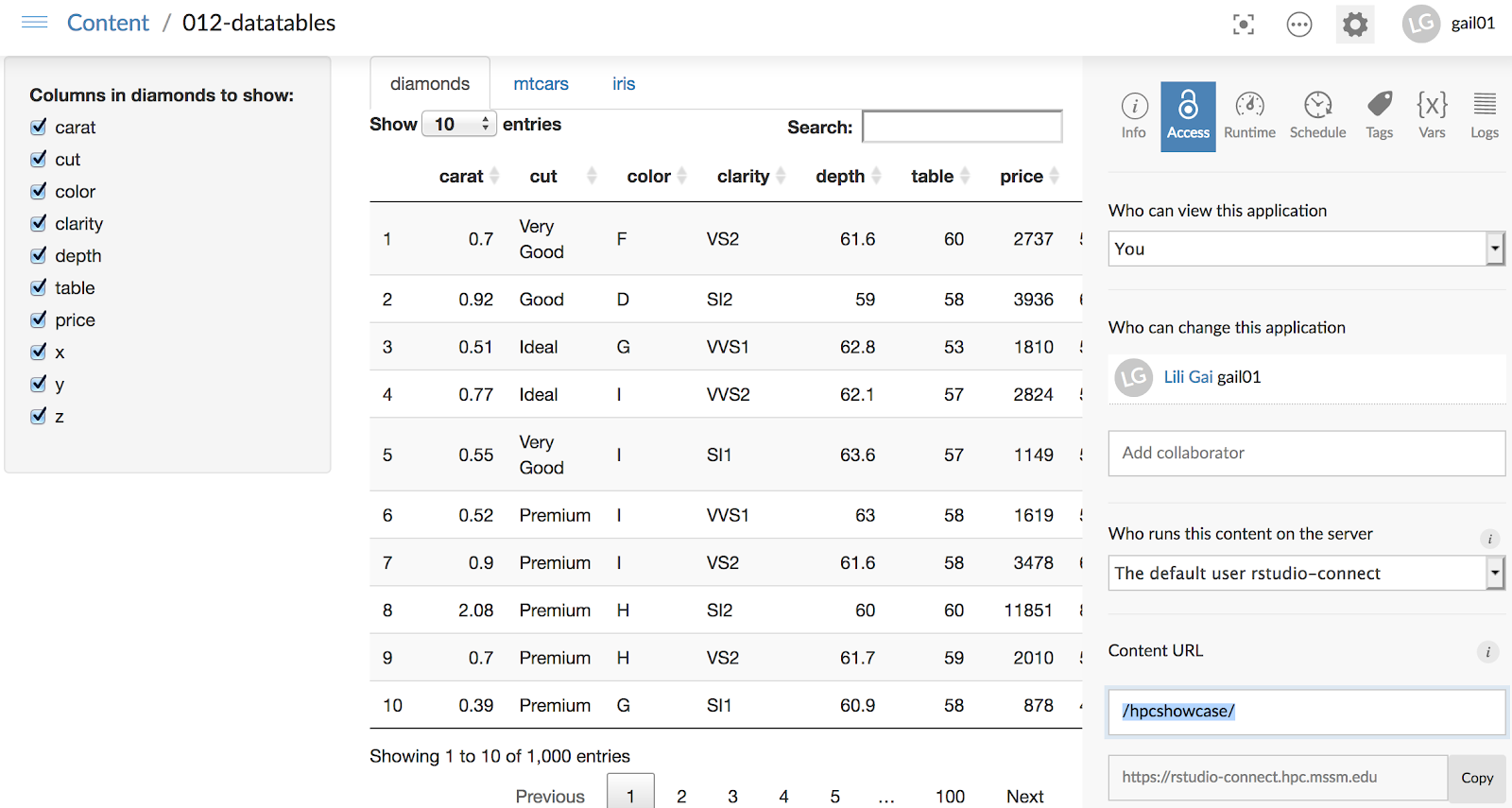
9. You can manage access and other metrics in the dashboard. Tip: you can modify your Content URL as shown in the screenshot (bottom right corner), and copy the URL for sharing. For more on how to manage your application in the dashboard, please follow Rstudio connect user guide located here.