TCGA – The Cancer Genome Atlas Program
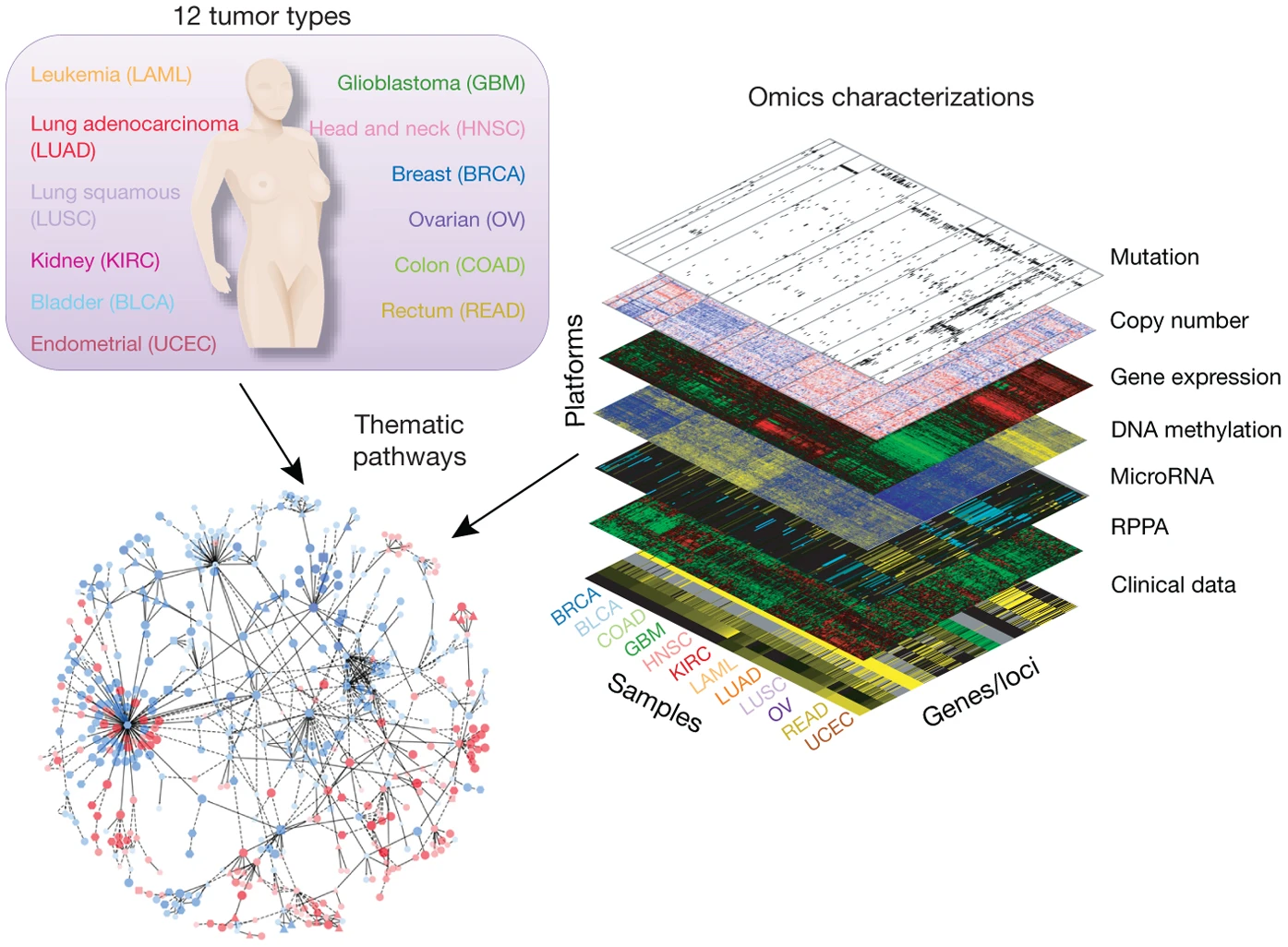
The Cancer Genome Atlas(TCGA) is a landmark cancer genomics program, that molecularly characterized over 20,000 primary cancer and matched normal samples spanning 33 cancer types. The program is a joint effort between National Cancer Institute and the National Human Genome Research Institute since 2016.
Currently, two versions are hosted on the Data Ark. Version 31.0 and version 32.0. The gene model used as a reference across TCGA has been updated from GENCODE 22(GRC37/hg19)—version 31 to GENCODE 36 (GRCh38/hg38)–version32. To learn more about the data sets from a different version, find the data release notes here
All the TCGA data sets downloaded belong to the “open-access” category and were obtained from the Genomic Data Commons Data Portal.
TCGA Processed Data Sets
TCGA Processed data sets
The TCGA RNA-seq counts files have been processed into 33 folders.
To use this data, NO DUA form is required, you can access the data at the following path on Minerva – /sc/arion/projects/data-ark/Public_Unrestricted/gnomAD or you can load module $ module load dataark to see the path variables.
Data Ark Data Sets
Public data sets (unrestricted)
Public data sets (restricted)
Mount Sinai generated data (unrestricted)
Mount Sinai generated data (restricted)
Data access
