Scientific Computing and Data / Research Data Services / Data Ark: Data Commons / Reference Genome
Reference Genome and Annotation
Overview
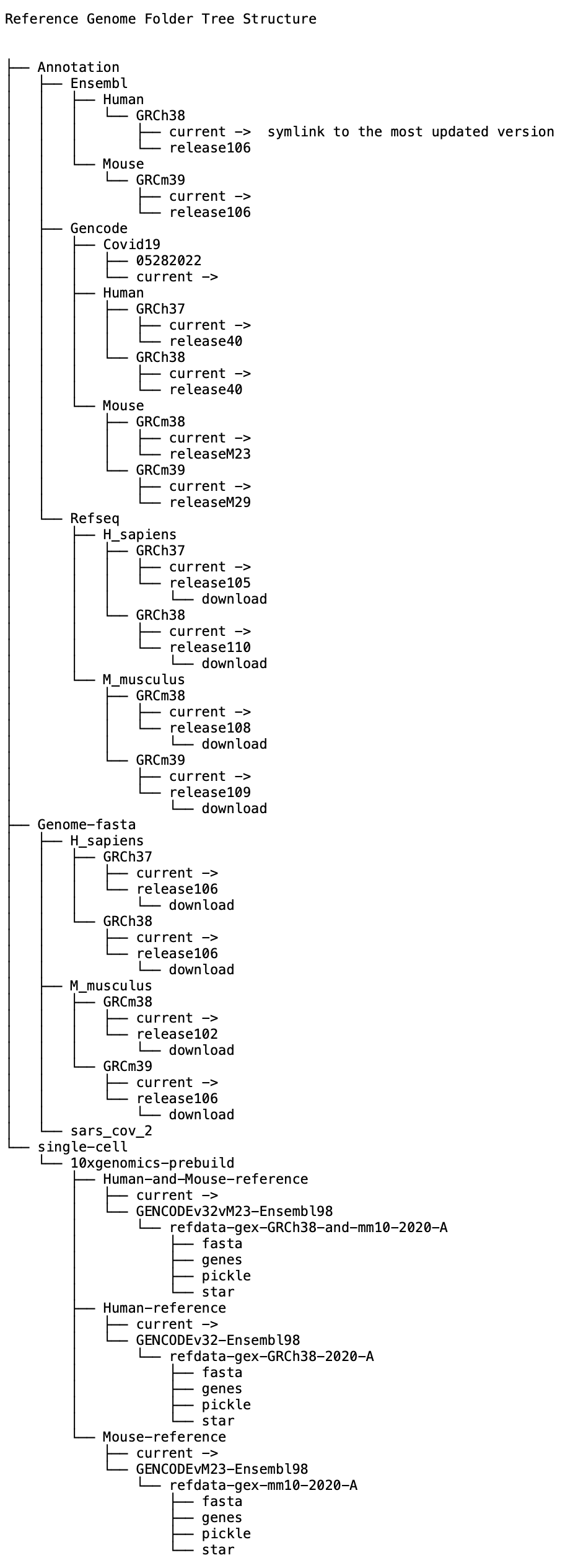
Data Ark is building an accessible reference genome resource folder. The folder covers the most frequently used reference genome (.fasta file) and annotation files (.tdf file).
The Genome files are downloaded from Ensemble Release 106, and the annotation files are downloaded from Ensemble, Refseq, and Gencode.
Between Gencode and Ensemble, the gene annotation is the same in both files. The only exception is that the genes which are common to the human chromosome X and Y PAR regions can be found twice in the GENCODE GTF, while they are shown only for chromosome X in the Ensembl file.
In general, the GENCODE/Ensemble annotations are more comprehensive–contain more exons, have greater genomic coverage, and capture many more variants than RefSeq in both genome and exome datasets, you can find more information through this paper link.
For the purpose of version control, we have a “current” version which is a symlink that always points to the most updated version of the file.
Access
Effective from January 22, 2024, you must read, agree and sign the Data Use Agreement (you must be logged in through the Mount Sinai campus network or secure remote VPN). Access is granted within 24 hours, and on Minerva, you can load module $ module load dataark to see the path variables.
Data Ark Data Sets
Please visit the Data Ark Data Set webpage to explore other data sets.
