gnomAD – The Genome Aggregation Database
Overview
The Genome Aggregation Database (gnomAD) is a resource developed by an international coalition of investigators, with the goal of aggregating and harmonizing both exome and genome sequencing data from a wide variety of large-scale sequencing projects, and making summary data available for the wider scientific community. Click here to learn more about gnomAD.
Currently, two data sets are hosted on the Data Ark.
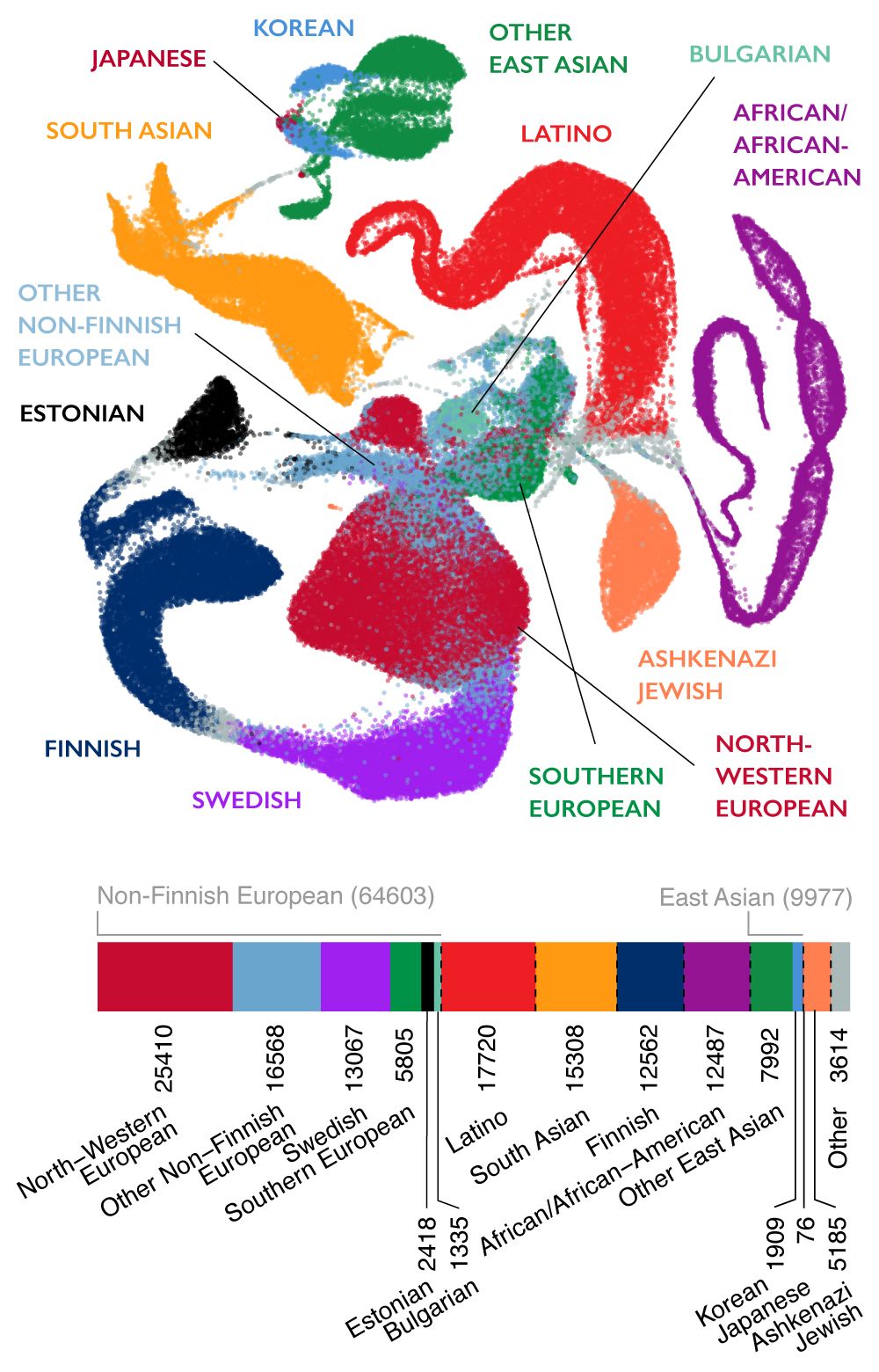
v2.1.1 data set (GRCh37/hg19) provided spans 125,748 exome sequences and 15,708 whole-genome sequences from unrelated individuals sequenced as part of various disease-specific and population genetic studies. gnomAD v2.1.1 is preferable over v3 for interpreting coding variants. The v3.1 data set (GRCh38) spans 76,156 genomes, providing more data for noncoding regions or coding regions not covered well in exomes, such as regions with high GC content or regions not targeted with exome capture. The v.3.1 data set also included genomes from the Human Genome Diversity Project (HGDP) and the 1000 Genome Project (1KG ).
Access
Effective from January 22, 2024, you must read, agree and sign the Data Use Agreement (you must be logged in through the Mount Sinai campus network or secure remote VPN). Access is granted within 24 hours, and on Minerva, you can load module $ module load dataark to see the path variables.
Data Ark Data Sets
Please visit the Data Ark Data Set webpage to explore other data sets.
