Scientific Computing and Data / AIR·MS (AI Ready Mount Sinai) / Getting Started
AIR·MS Data Modalities
The AIR·MS platform proudly features the following Mount Sinai and public datasets. Our team is dedicated to continuously expanding our database with additional data modalities, striving to build a comprehensive, multi-modal research resource. To help you get started with the data, we offer Quick Start guides available here.
Mount Sinai Datasets
|
|
|
|
||||||||
|
|
|
|
||||||||
|
Public Datasets
- Medicare Claims Synthetic Public Use Files (SynPUFs)

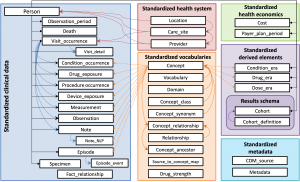
Mount Sinai Data Warehouse (MSDW) in OMOP format
The MSDW dataset leverages the OMOP Common Data Model. The data is comprised of clinical data extracted from Mount Sinai’s Epic Caboodle database and other ancillary systems.
We offer both an identifiable and a de-identified version of the MSDW dataset in AIR·MS
MSDW OMOP identifiable (PHI) with extended attributes
Current Status
Schema: CDMPHI
Data snapshot from 04/16/2025
Unique patients: 12,107,621
MSDW OMOP de-identified (de-id)
Current Status
Schema: CDMDEID
Data snapshot from 09/26/2023
Unique patients: 10,934,180
| Table | Record Count |
|---|---|
| CARE_SITE | 106,699 |
| CDC_RACE_ETHNICITY_XTN | 967 |
| CONCEPT | 11,337,020 |
| CONCEPT_ANCESTOR | 101,072,304 |
| CONCEPT_CLASS | 453 |
| CONCEPT_RELATIONSHIP | 170,586,066 |
| CONCEPT_SYNONYM | 5,190,521 |
| CONDITION_OCCURRENCE | 206,228,288 |
| DEATH | 49,797 |
| DOMAIN | 50 |
| DRUG_EXPOSURE | 213,932,157 |
| DRUG_STRENGTH | 3,003,619 |
| FACT_RELATIONSHIP | 161,135,148 |
| LOCATION | 13,394,927 |
| MEASUREMENT | 1,875,881,345 |
| NOTE | 242,610,978 |
| OBSERVATION | 516,585,500 |
| OBSERVATION_PERIOD | 12,147,842 |
| PERSON | 12,107,621 |
| PROCEDURE_OCCURRENCE | 312,011,901 |
| PROVIDER | 1,363,962 |
| PROVIDER_ATTRIBUTE_XTN | 783,560 |
| RELATIONSHIP | 730 |
| VISIT_OCCURRENCE | 198,943,396 |
| VOCABULARY | 254 |
Note
Some of the standard OMOP tables contain extension fields (starting with the prefix ‘XTN’) which contain data outside of the OMOP standard data model. Many of these XTN attributes are based on data derived directly from EPIC (i.e. codes used in EPIC rather than the standardized OMOP codes), or attributes not currently contained in the OMOP standard.
| Table | Record Count |
| MEASUREMENT | 1,330,106,439 |
| OBSERVATION | 274,496,232 |
| PROCEDURE_OCCURRENCE | 192,252,154 |
| DRUG_EXPOSURE | 149,311,961 |
| CONCEPT_RELATIONSHIP | 143,179,970 |
| NOTE | 137,886,581 |
| CONDITION_OCCURRENCE | 122,966,185 |
| VISIT_OCCURRENCE | 118,936,818 |
| FACT_RELATIONSHIP | 118,710,809 |
| CONCEPT_ANCESTOR | 94,315,531 |
| CONCEPT_SYNONYM | 11,899,964 |
| OBSERVATION_PERIOD | 10,955,831 |
| PERSON | 10,934,180 |
| CONCEPT | 10,302,430 |
| DRUG_STRENGTH | 2,994,169 |
| LOCATION | 1,321,549 |
| PROVIDER | 1,307,112 |
| CARE_SITE | 301,467 |
| DEATH | 3,213 |
| RELATIONSHIP | 692 |
| CONCEPT_CLASS | 437 |
| VOCABULARY | 251 |
| DOMAIN | 50 |

Clinical Notes
Clinical notes in the form of unstructured data (progress notes, telephone encounters, nursing notes, procedures, etc.) extracted from the MSDW OMOP identifiable dataset have been loaded to AIR.MS and enabled for search using SAP HANA’s in-memory full-text search capabilities. This feature empowers the researcher to build patient cohorts based on terms contained in unstructured reports in seconds or even milliseconds! The researcher can further filter based on note type or an array of other clinical attributes.
Tip: See the Getting Started Guides for examples on how to perform search using python!
Current Status
Schema: CDMPHI
Data snapshot from 04/16/2025
Number of indexed notes: 242,610,978

Pathology (metadata)
The Pathology metadata aids researchers in the field of Computational Pathology. Researchers are able to query the metadata in combination with other linked data modalities to build a patient cohort, subsequently apply quantitative methods for the analysis of digital microscopy slides and relating the resulting statistical descriptors to patient outcomes.
We are also working on making the digital slides available to researchers on Minerva HPC.
Current Status
Data Source: Powerpath
Schema: CDMPATHOLOGY
Data snapshot from 01/08/2025
Unique patients: 3,528,719
Note
Pathology reports are now available in AIR·MS and can be found in table ACC_RESULTS. The reports are broken up into sections (clinical history, final diagnosis, SNOMED coding, etc.) that can be identified by column PATH_RPT_HEADING_NAME. If you are looking for the full report, you can combine all records for with the same ACCESSION_2_ID into a single output. The column containing the free-text (ACC_RESULTS_FINDING) has been enabled with SAP HANA full-text search capabilities. Examples of how to use HANA full-text search are available in this tutorial notebook.
The following tables and attributes are available in AIR·MS:
ACCESSION
Number of records: 7,107,464
| Column Name | Comments |
|---|---|
| ACC_CATG | Case category |
| ACC_PROCESS_STEP_COMPLETED_DATE | Case finalize date / status update datetime |
| ACCESSION_2_ID | PowerPath unique case ID |
| ACCESSION_NO | Case number |
| BIRTH_DATE | Patient date of birth |
| CREATED_DATE | Case creation date |
| CURRENT_STATUS_ID | PowerPath unique identifier for a case status |
| FACILITY_CODE | Facility code |
| FACILITY_ID | PowerPath ID for the facility associated with the accession |
| FACILITY_NAME | Facility name |
| IMPORTED_CASE | One-character \”Y\” or \”N\” code indicating if the case was imported into PowerPath |
| LAST_UPDATE_DATETIME | Case finalize date / status update datetime |
| MED_REC_NO | Medical record number (EPIC MRN) |
| MRN_FACILITY_CODE | PowerPath ID for the facility that assigned the MRN |
| MRN_FACILITY_DESCRIPTION | Name of the facility that assigned the MRN |
| ORDER_NUMBER | Order number from the ordering system |
| PATIENT_AGE | The patient’s age on the case creation date |
| PATIENT_ID | PowerPath patient ID |
| PERSONNEL_2_FULL_NAME | Name of the pathologist who finalized the accession |
| PERSONNEL_2_ID | PowerPath ID for the pathologist who finalized the accession |
| PROCESS_STEP_DESCRIPTION | Case status name / description |
| VISIT_NUMBER | Encounter identifier |
ACC_ICD
Number of records: 3,548,815
| Column Name | Comments |
|---|---|
| ACC_ICD9_ID | PowerPath surrogate unique identifier for an ICD-10 code assigned to a case |
| ACCESSION_2_ID | PowerPath unique case ID |
| LAST_UPDATE_DATETIME | Case finalize date / status update datetime |
| MEDICAL_CODE | ICD-10 code assigned for billing |
| MEDICAL_CODE_ID | PowerPath surrogate unique identifier for an ICD-10 code |
ACC_SLIDE
Number of records: 14,241,494
| Column Name | Comments |
|---|---|
| ACC_BLOCK_ID | PowerPath unique block ID |
| ACC_BLOCK_LABEL | Specimen block identifier |
| ACC_PROCESS_STEP_COMPLETED_DATE | Case finalize date / status update datetime |
| ACC_SLIDE_ID | PowerPath unique slide ID |
| ACC_SPECIMEN_DESCRIPTION | Specimen source description |
| ACC_SPECIMEN_ID | PowerPath unique specimen ID |
| ACCESSION_2_ID | PowerPath unique case ID |
| BIOPSY | Boolean flag for 1 = biopsy, 0 = non-biopsy |
| COLLECTION_DATE | Specimen collection date |
| CONSULT_LABEL | Optional free text for slides of type \”consult\” |
| LAB_PROCEDURE_CODE | The procedure code |
| LAB_PROCEDURE_DESCRIPTION | The procedure description |
| LAB_PROCEDURE_ID | PowerPath unique procedure identifier |
| LAST_UPDATE_DATETIME | Case finalize date / status update datetime |
| RECV_DATE | Specimen received date |
| SLIDE_LABEL | Derived unique (business key) identifier for each slide |
| SLIDE_NO | Ordinal number of the slide from the specimen & block |
| SLIDE_TYPE | Whether the slide is stained, unstained or antibody/IHC |
| SOURCE_MATERIAL_LABEL | Derived unique (business key) identifier for each slide’s source specimen and block |
| SOURCE_REC_TYPE | Where the slide came from, either specimen or block |
| SPECIMEN_CATEGORY_ID | PowerPath specimen category ID |
| SPECIMEN_CATEGORY_NAME | The specimen category name |
| SPECIMEN_GROUPS_CODE | Specimen specialty code |
| SPECIMEN_GROUPS_ID | PowerPath specimen specialty ID |
| SPECIMEN_LABEL | Specimen identifier |
| TYPE | Whether the slide is consult or not consult |
ACC_RESULTS
Number of records: 31,193,765
| Column Name | Comments |
|---|---|
| ACC_RESULTS_FINDING | The text of the report section |
| ACC_RESULTS_ID | PowerPath unique identifier for each report section |
| ACC_RESULTS_REC_ID | sort order of result section on RTF |
| ACCESSION_2_ID | PowerPath unique case ID |
| LAST_UPDATE_DATETIME | Case finalize date / status update datetime |
| PATH_RPT_HEADING_ID | PowerPath result section heading on RTF |
| PATH_RPT_HEADING_NAME | PowerPath result section heading name |
ACC_SLIDE_IMAGESERVER
Number of records: 2,564,239
| Column Name | Comments |
|---|---|
| ACC_SLIDE_ID | PowerPath unique slide ID |
| ACC_SLIDE_IMAGESERVER_DESCRIPTION | The name of the Philips iSyntax slide image file |
| ACC_SLIDE_IMAGESERVER_ID | PowerPath unique identifier for a slide image |
| INTERNAL_SLIDE_ID | Identifier for the slide, also known as the \”barcode\” ID |
| LAST_UPDATE_DATETIME | Case finalize date / status update datetime |
| SCAN_DATE | The date on which the slide image was digitized |

Radiology (metadata)
AIR·MS now features radiology metadata extracted from the Mount Sinai IRW 2.0 XNAT system (via an MSDW data pipeline). This data set is comprised of detailed DICOM (Digital Imaging and Communications in Medicine) tags associated with the medical images.
These tags provide essential metadata, including patient information, imaging parameters, equipment details, and procedural context, ensuring a comprehensive understanding of each radiological study.
By integrating this metadata, we enable researchers to gain deeper insights into the imaging data, facilitating advanced analyses and fostering innovations in medical imaging research.
Current Status
Schema: CDMRADIOLOGY
Data snapshot from 8/16/2024
Unique patients: 2,057,482
The following tables and attributes are available in AIR·MS:
Number of records: 66,993,826
| RADIOLOGY_METADATA |
| ID |
| PATIENT_ID |
| SERIES_INSTANCE_UID |
| STUDY_INSTANCE_UID |
| ETL_RECORD_UPDATE_DATETIME |
Number of records: 7,745,407,087
| RADIOLOGY_DICOM_DATA |
| ID |
| RADIOLOGY_METADATA_ID |
| DICOM_TAGS |
| TAG_VALUE_REPRESENTATION |
| TAG_VALUE” NCLOB MEMORY |
| TAG_XTN_PATIENT_EPIC_MRN |
| TAG_PATIENT_NAME |
| SERIES_INSTANCE_UID |
| STUDY_INSTANCE_UID |
| ETL_RECORD_UPDATE_DATETIME |
The current lack of diversity in genomic research data is hindering what we can learn about health and potential treatments in our global population. By enhancing the diversity of people participating in genomic research, we can advance our knowledge and discovery of human genetics for all populations. To that end, The Charles Bronfman Institute for Personalized Medicine is spearheading the effort to carry out the genetic sequencing of one million Mount Sinai patients within the next five years. This initiative, one of the largest such sequencing projects of its kind, will integrate health and research data at Mount Sinai to promote discoveries that will directly benefit our patient population.
Access to the BioMe Biobank and Mount Sinai Million Biobank on HPC can be requested via the CBIPM Data and Specimen Inquiry Form
The following tables and attributes are available in AIR·MS:
Mount Sinai Million / BioMe identifiable (PHI)
Current Status
Schema: CDMMSM
Data snapshot from 06/09/2025
Unique patients: 279,885
Mount Sinai Million / BioMe de-identified (de-id)
Current Status
Schema: CDMMSMDEID
Data snapshot from 06/09/2025
Unique patients: 279,885
| PATIENT | Comments |
| ID | Internal ID for linking to other tables within the dataset |
| MRN | Medical Record Number (EPIC MRN – only accessible under regulatory approval) |
| MASKED_MRN | De-Identifier for combined BioMe Biobank set with Regeneron and Sema4 data |
| RGN_ID | De-identifier for first Regeneron batch regarding BioMe Biobank |
| SEMA4_ID | De-identifier for Sema4, a subset of Masked MRN ID |
| MSM_ID | De-identifier for Mount Sinai Million Biobank, a combined setoff RGN_ID and new MSM ID |
| MILLION_ID | Indicator for all consented patients with and without genomic data |
| AIR_CREATED_AT | Record creation in AIR·MS |
| AIR_UPDATED_AT | Record updated in AIR·MS |
| PATIENT | Comments |
| ID | Internal ID for linking to other tables within the dataset |
| MASKED_MRN | De-Identifier for combined BioMe Biobank set with Regeneron and Sema4 data |
| RGN_ID | De-identifier for first Regeneron batch regarding BioMe Biobank |
| SEMA4_ID | De-identifier for Sema4, a subset of Masked MRN ID |
| MSM_ID | De-identifier for Mount Sinai Million Biobank, a combined setoff RGN_ID and new MSM ID |
| MILLION_ID | Indicator for all consented patients with and without genomic data |
| AIR_CREATED_AT | Record creation in AIR·MS |
| AIR_UPDATED_AT | Record updated in AIR·MS |

Electrocardiogram (ECG)
Current Status
Data Source: GE HealthCare MUSE Cardiology Information System
Schema: CDMECG
Data snapshot from: 04/10/2021
Unique patients: 1,961,254
The following tables and attributes are available in AIR·MS:
Number of records: 9,275,130
| PATIENT_DEMOGRAPHICS |
| PATIENT_DEMOGRAPHICS_ID (X) |
| FILE_ENTRY_ID |
| PATIENT_ID |
| PATIENTAGE |
| AGEUNITS |
| DATEOFBIRTH |
| GENDER |
| RACE |
| PATIENTLASTNAME |
| PATIENTFIRSTNAME |
Number of records: 9,168,266
| DIAGNOSIS |
| DIAGNOSIS_ID (X) |
| FILE_ENTRY_ID |
| MODALITY |
| DIAGNOSISSTATEMENT |
Number of records: 73,631,055
| LEAD_DATA |
| LEAD_DATA_ID (X) |
| FILE_ENTRY_ID |
| LEADBYTECOUNTTOTAL |
| LEADTIMEOFFSET |
| LEADSAMPLECOUNTTOTAL |
| LEADAMPLITUDEUNITSPERBIT |
| LEADAMPLITUDEUNITS |
| LEADHIGHLIMIT |
| LEADLOWLIMIT |
| LEADID |
| LEADOFFSETFIRSTSAMPLE |
| FIRSTSAMPLEBASELINE |
| LEADSAMPLESIZE |
| LEADOFF |
| BASELINESWAY |
| LEADDATACRC32 |
| WAVEFORMDATA |
Number of records: 9,610,935
| ECG_FILES |
| FILE_ENTRY_ID (X) |
| FILE_NAME |
| FILE_PATH |
| FILE_HASH |
| FILE_SIZE_BYTES |
| ACQUISITION_DATE |
| ACQUISITION_TIME |
| PROCESSING_STATUS |
| STATUS_CODE |
| NOTES_AND_COMMENTS |
| FILE_TIMESTAMP |
| AIR_CREATED_AT |
| AIR_UPDATED_AT |
| JSON_STATUS |
Number of records: 9,168,230
| MUSE_INFO |
| MUSEVERSION |
| FILE_ENTRY_ID |
Number of records: 7,757,472
| ORDER_INFO |
| ORDER_INFO_ID (X) |
| FILE_ENTRY_ID |
| HISACCOUNTNUMBER |
| ORDERTIME |
| ADMITTIME |
| ADMITDATE |
| HISLOCATION |
| BED |
| ATTENDINGMDHISID |
| ATTENDINGMDLASTNAME |
| ATTENDINGMDFIRSTNAME |
| ALTERNATEVISITID |
| HISDISPOSITION |
| ADMITSOURCE |
| PRIMARYDIAGNOSTICCODE |
| SERVICINGFACILITY |
| ADMITTINGMDHISID |
| ADMITTINGMDLASTNAME |
| ADMITTINGMDFIRSTNAME |
| CONSULTINGMDID |
| REFERRINGMDHISID |
| HOSPITALSERVICE |
| ADMISSIONTYPE |
Number of records: 9,164,220
| ORIGINAL_DIAGNOSIS |
| ORIGINAL_DIAGNOSIS_ID (X) |
| FILE_ENTRY_ID |
| MODALITY |
| DIAGNOSISSTATEMENT |
Number of records: 9,168,044
| ORIGINAL_RESTING_ECG_MEASUREMENTS |
| ORIGINAL_RESTING_ECG_MEASUREMENTS_ID (X) |
| VENTRICULARRATE |
| ATRIALRATE |
| PRINTERVAL |
| QRSDURATION |
| QTINTERVAL |
| QTCORRECTED |
| PAXIS |
| RAXIS |
| TAXIS |
| QRSCOUNT |
| QONSET |
| QOFFSET |
| PONSET |
| POFFSET |
| TOFFSET |
| ECGSAMPLEBASE |
| ECGSAMPLEEXPONENT |
| QTCFREDERICA |
Number of records: 3,613,372
| PHARMA_DATA |
| PHARMA_DATA_ID (X) |
| PHARMARRINTERVAL |
| PHARMAUNIQUEECGID |
| PHARMAPPINTERVAL |
| PHARMACARTID |
| FILE_ENTRY_ID |
Number of records: 9,649,712
| QRS_TIMES_TYPES |
| GLOBALRR |
| QTRGGR |
| FILE_ENTRY_ID |
Number of records: 9,657,368
| RESTING_ECG |
| RESTING_ECG_ID (X) |
| FILE_ENTRY_ID |
| PATIENT_ID |
| ACQUISITIONDATE |
| ACQUISITIONTIME |
| STATUS |
Number of records: 9,167,778
| RESTING_ECG_MEASUREMENTS |
| RESTING_ECG_MEASUREMENTS_ID (X) |
| FILE_ENTRY_ID |
| VENTRICULARRATE |
| ATRIALRATE |
| PRINTERVAL |
| QRSDURATION |
| QTINTERVAL |
| QTCORRECTED |
| PAXIS |
| RAXIS |
| TAXIS |
| QRSCOUNT |
| QONSET |
| QOFFSET |
| PONSET |
| POFFSET |
| TOFFSET |
| ECGSAMPLEBASE |
| ECGSAMPLEEXPONENT |
| QTCFREDERICA |
Number of records: 9,654,325
| TEST_DEMOGRAPHICS |
| TEST_DEMOGRAPHICS_ID (X) |
| FILE_ENTRY_ID |
| DATATYPE |
| SITE |
| SITENAME |
| ACQUISITIONDEVICE |
| STATUS |
| EDITLISTSTATUS |
| PRIORITY |
| LOCATION |
| LOCATIONNAME |
| ROOMID |
| ACQUISITIONTIME |
| ACQUISITIONDATE |
| CARTNUMBER |
| ACQUISITIONSOFTWAREVERSION |
| ANALYSISSOFTWAREVERSION |
| EDITTIME |
| EDITDATE |
| EDITORID |
| REFERRINGMDLASTNAME |
| REFERRINGMDFIRSTNAME |
| ACQUISITIONTECHLASTNAME |
| EDITORLASTNAME |
| EDITORFIRSTNAME |
| SECONDARYID |
| HISSTATUS |
/482268916-Echocardiogram-image-scaled.jpeg)
Cardiovascular Imaging (metadata)
AIR·MS contains DICOM metadata tags for cardiovascular imaging studies performed in the Mount Sinai Health System that are contained within the Softlink cardiovascular PACS system. This does not include radiology data contained within the radiology PACS systems. The following common modalities include: US, CT, XA, NM, MR, IVUS. These modalities include,
among others, echocardiographic ultrasound, vascular ultrasound, and angiographic data. These tags are linked to DICOM files by ECHO_METADATA.IMAGE_FILE_PATH in a repository on the Minerva cluster. Note that the schema name CDMECHO is a misnomer – this catalog
contains much more than echocardiogram data.
The catalog contains every public DICOM metadata tag (dictionaries are publicly available, see https://dicom.innolitics.com/ciods for instance). The
Tag is under ECHO_TAGS_DATA.TAGS and is labeled by its Name not its hexadecimal number for ease of readability (see
https://www.dicomlibrary.com/dicom/dicom-tags/ for dictionary of public tags and name). The value of the tag is under ECHO_TAGS_DATA.TAG_VALUE.
Notes:
-
Aas of late August 2025 there are known significant gaps in data availability around year 2019 and starting in mid 2023 and onward. The catalog ends in late 2023. Mechanisms for capturing missing data and updating the catalog moving forward are underway.
-
There will be some duplicated data within the archive (i.e. 2 identical studies may be in 2 separate paths)
-
There is a known bug in the way the value representation of “Person Name” is stored. It is stored as a list of single characters rather than a
string (i.e. the name Smith is stored as [S,m,i,t,h])
Current Status
Schema: CDMECHO
Data snapshot from: 10/17/2023
Unique patients: 885,957
The following tables and attributes are available in AIR·MS:
Number of records: 268,682,931
| ECHO_METADATA |
| ID |
| FILE_ENTRY_ID |
| PATIENT_ID |
| SERIES_INSTANCE_UID |
| STUDY_INSTANCE_UID |
| SOP_INSTANCE_UID |
| IMAGE_FILE_PATH |
| AIR_CREATED_AT |
| AIR_UPDATED_AT |
Number of records: 21,089,507,788
| ECHO_TAGS_DATA |
| ID |
| FILE_ENTRY_ID |
| SERIES_INSTANCE_UID |
| STUDY_INSTANCE_UID |
| SOP_INSTANCE_UID |
| TAGS |
| TAG_VALUE |
| AIR_CREATED_AT |
| AIR_UPDATED_AT |
/3616198284.png)
Synthetic Public Use File (DE-SynPUF)
The SYNPUF (Synthetic Public Use Files) dataset, provided by the Centers for Medicare & Medicaid Services (CMS), offers a synthetic version of Medicare claims data from the years 2008 to 2010. This dataset is meticulously designed to maintain the statistical properties and relationships present in the original data while ensuring that no actual patient information is disclosed, thereby safeguarding privacy.
SYNPUF includes a comprehensive array of variables such as beneficiary demographics, chronic conditions, hospital and outpatient claims, and prescription drug events, making it an invaluable resource for researchers and data scientists. It serves as an exemplary tool for developing and testing healthcare models, algorithms, and applications without the constraints associated with sensitive real-world data.
The SYNPUF dataset in AIR·MS utilizes the OMOP Common Data Model, aligned with other clinical data sets available on the platform. Since SYNPUF data does not require an approved IRB, you can easily get onboarded and start building ML models!
Current Status
Schema: CDMSYNPUF
Number of patients: 2,326,856
Number of observations: 37,531,051
Number of measurements: 72,387,791

Work in Progress
We are constantly integrating new data to AIR·MS. The following data modalities are currently being worked on:
-
Electroencephalogram (EEG)
-
Endoscopy & colonoscopy reports
-
Bedmaster
-
Radiology images and reports