Our curated resources centralize detailed information about genes, proteins, drugs, and their functions and interactions. By organizing and serving this knowledge, the resources support hypothesis generation and data-driven exploration. Using ML/AI, these resources also provide predictions for the functions of all human genes, as well as targets and side effects for preclinical compounds.
Browse through the following categories to explore the various original resources we have developed:
Most Popular | Recently Published | Enrichment Analysis | Drug and Target Discovery | Computational Platforms and Workflow Engines | Gene and Drug Pages | lncRNAs | Data Portals | Data Visualization Components | Deprecated
GeneRanger
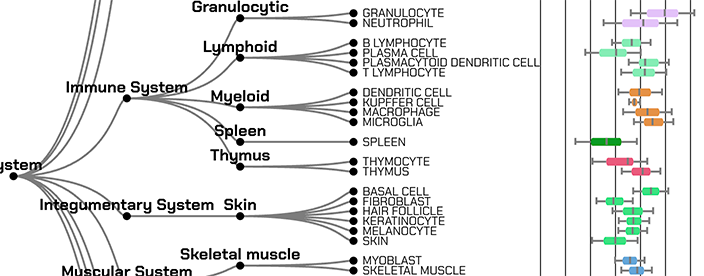
Expression of Human Genes and Proteins across Human Cell Types, Tissues, and Cell Lines across Multiple Atlases
GeneRanger is a web-server application that provides access to processed data about the expression of human genes and proteins across human cell types, tissues, and cell lines from several atlases.
PMID: 37166966
PrismEXP
Prediction of Gene Insights from Stratified Mammalian Gene Co-Expression
Using uniformly aligned data from ARCHS4, we apply PrismEXP to predict a wide variety of gene annotations including pathway membership, Gene Ontology terms, as well as human and mouse phenotypes. Predictions made with PrismEXP outperform predictions made with the global cross-tissue co-expression correlation matrix approach on all tested domains, and training using one annotation domain can be used to predict annotations in other domains. The PrismEXP web-based application, with pre-computed PrismEXP predictions, is available from: https://maayanlab.cloud/prismexp.
PrismEXP Appyter
PrismEXP Python package
PMID: 36874981
GDLPA
Gene and Drug Landing Page Aggregator
Gene and Drug Landing Page Aggregator (GDLPA) has links to 53 gene, 18 variant and 19 drug repositories that provide direct links to gene and drug landing pages. You can search by gene or drug name and then choose the sites that contain knowledge about your gene or drug of interest.
PMID: 35368424
ARCHS4
All RNA-seq and ChIP-seq Signature Search Space
ARCHS4 provides access to gene counts from HiSeq 2000 and HiSeq 2500 platforms for human and mouse experiments from GEO and SRA. The website enables downloading of the data in H5 format for programmatic access as well as a 3-dimensional view of the sample and gene spaces. Search features allow browsing of the data by meta data annotation, ability to submit your own up and down gene sets, and explore matching samples enriched for annotated gene sets. Selected sample sets can be downloaded into a tab separated text file through auto-generated R scripts for further analysis. Reads are aligned with Kallisto using a custom cloud computing platform. Human samples are aligned against the GRCh38 human reference genome, and mouse samples against the GRCm38 mouse reference genome.
PMID: 29636450
Geneshot
Submit Biomedical Terms to Receive Ranked Lists of Relevant Genes
Enables researchers to enter arbitrary search terms, to receive ranked lists of genes relevant to the search terms. Returned ranked gene lists contain genes that were previously published in association with the search terms, as well as genes predicted to be associated with the terms based on data integration from multiple sources. The search results are presented with interactive visualizations.
PMID: 31114885
DrugShot
Querying Biomedical Search Terms to Retrieve Prioritized Lists of Small Molecules
DrugShot is a web-based server application and an Appyter that enables users to enter any biomedical search term into a simple input form to receive ranked lists of drugs and other small molecules based on their relevance to the search term.
PMID: 35183110
Drugmonizome
Search Engine for Querying Annotated Sets of Drugs and Small Molecules
A database with a search engine for querying annotated sets of drugs and small molecules for performing drug set enrichment analysis. Utilizing the data within Drugmonizome, we also developed Drugmonizome-ML which enables users to construct customized machine learning pipelines using the drug set libraries from Drugmonizome. To demonstrate the utility of Drugmonizome, drug sets from 12 independent SARS-CoV-2 in vitro screens were subjected to consensus enrichment analysis. Despite the low overlap among these 12 independent in vitro screens, we identified common biological processes critical for blocking viral replication. To demonstrate Drugmonizome-ML, we constructed a machine learning pipeline to predict whether approved and preclinical drugs may induce peripheral neuropathy as a potential side effect.
PMID: 33787872