Avi Ma’ayan’s Publications on PubMed | Google Scholar | ResearchGate
Featured Publication
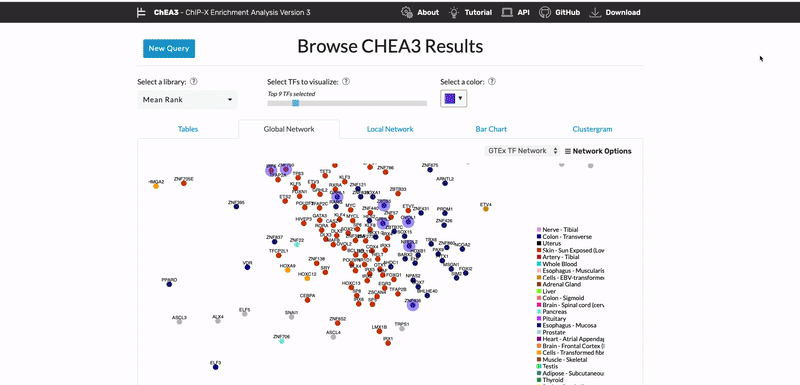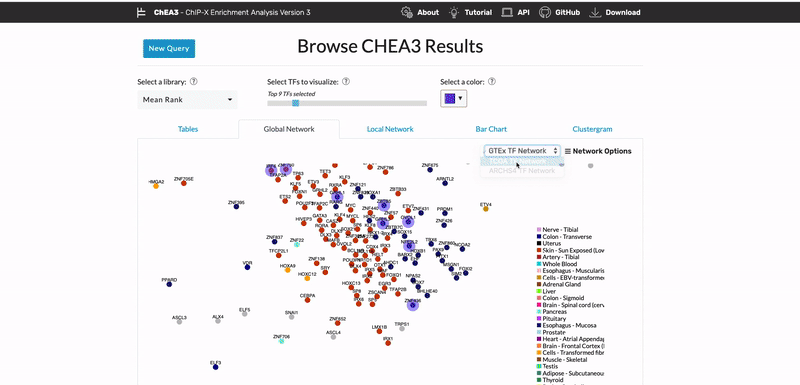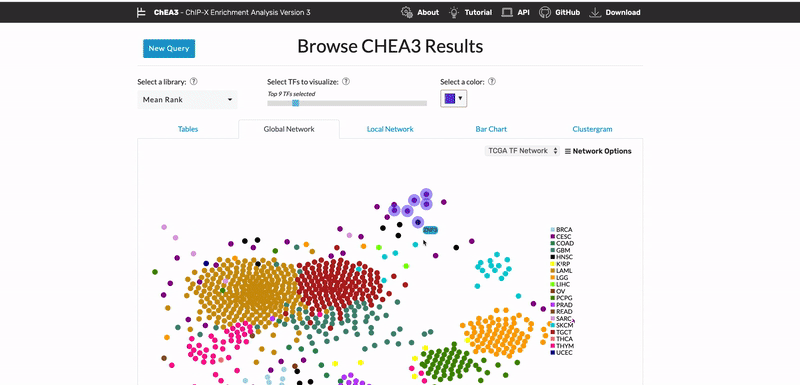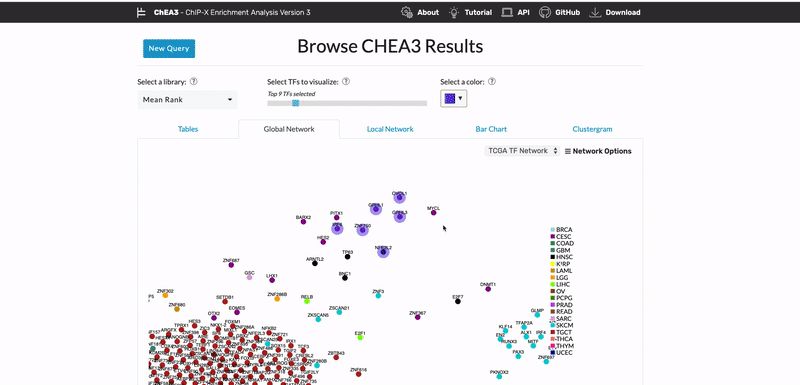
ChEA3: transcription factor enrichment analysis by orthogonal omics integration
2026
Evangelista JE, Clarke DJB, Byrd AI, Srinivasan S, Srinivasan S, Maurya MR, Jenkins SL, Diamant I, Sanchez E, Xie Z, Olaiya S, Kim H, Marino GB, Ahmed N, Ramachandran S, Subramaniam S, Ma’ayan A. The CFDE Workbench: Integrating metadata and processed data from Common Fund Programs. Journal of Molecular Biology 169631
2025
Marino GB, Byrd AI, Ahmed N, Clarke DJB, Ma’ayan A. sc2DAT: Workflow for targeting tumor subpopulations of single cells. Bioinformatics Advances vbaf237 (2025)
Evangelista JE, Lutsky AD, Byrd AI, Clarke DJB, Prabhakaran A, Jenkins SL, Ma’ayan A. Creating an interactive web interface for networks stored in knowledge graph databases. Current Protocols 5(9):e70200 (2025) PMID: 40956607
Byrd AI, Evangelista JE, Lachmann A, Chung HY, Jenkins SL, Ma’ayan A. ChEA-KG: Human transcription factor regulatory network with a knowledge graph interactive user interface. bioRxiv 2025.08.09.669505 (2025) PMID: 40832173
Satpathy S, […], Ma’ayan A, Getz GA, Dhanasekaran SM, Robles AI, Chang GC, Yang PC, Yu SL, Chen HY, Nesvizhskii AI, Carr SA, Mani DR, Cieslik MP, Chen YJ, Gillette MA; Taiwan Cancer Moonshot Program; Clinical Proteomic Tumor Analysis Consortium. Integrative analysis of lung adenocarcinoma across diverse ethnicities and exposures. Cancer Cell 43(9):1731-1757.e17 (2025) PMID: 40749670
Clarke DJB, Marinp GB, Ma’ayan A. A gene set foundation model pre-trained on a massive collection of diverse gene sets. bioRxiv 2025.05.30.657124 (2025) PMID: 0501705
Marino GB, Evangelista JE, Clarke DJB, Ma’ayan A. L2S2: chemical perturbation and CRISPR KO LINCS L1000 signature search engine. Nucleic Acids Research gkaf373 (2025) PMID: 40308216
Marino GB, Olaiya S, Evangelista JE, Clarke DJB, Ma’ayan A. GeneSetCart: assembling, augmenting, combining, visualizing, and analyzing gene sets. GigaScience 14:giaf025 (2025) PMID: 40208796
Clarke DJB, Evangelista JE, Xie Z, Marino GB, Byrd AI, Maurya MR, Srinivasan S, Yu K, Petrosyan V, Roth ME, Milinkov M, King CH, Vora JK, Keeney J, Nemarich C, Khan W, Lachmann A, Ahmed N, Agris A, Pan J, Ramachandran S, Fahy E, Esquivel E, Mihajlovic A, Jevtic B, Milinovic V, Kim S, McNeely P, Wang T, Wenger E, Brown MA, Sickler A, Zhu Y, Jenkins SL, Blood PD, Taylor DM, Resnick AC, Mazumder R, Milosavljevic A, Subramaniam S, Ma’ayan A. Playbook workflow builder: Interactive construction of bioinformatics workflows. PLOS Computational Biology 21(4):e1012901 (2025) PMID: 40179105
Evangelista JE, Clarke DJB, Xie Z, Olaiya S, Kim H, Marino GB, Byrd A, Srinivasan S, Srinivasan S, Maurya MR, Jenkins SL, Lutsky AD, Sasaya L, Lachmann A, Ahmed N, Diamant I, Sanchez E, Ramachandran S, Subramaniam S, Ma’ayan A. The CFDE Workbench: integrating metadata and processed data from Common Fund programs. bioRxiv 2025.02.04.636535 (2025)
Evangelista JE, Ali-Nasser T, Malek LE, Xie X, Marino GB, Bester AC, Ma’ayan A. lncRNAlyzr: Enrichment analysis for lncRNA sets. Journal of Molecular Biology 168938 (2025) PMID: 40133794
2024
Diamant I, Clarke DJB, Evangelista JE, Lingam N, Ma’ayan A. Harmonizome 3.0: integrated knowledge about genes and proteins from diverse multi-omics resources. Nucleic Acids Research gkae1080 (2024) PMID: 39565209
Marino GB, Deng EZ, Clarke DJB, Diamant I, Resnick AC, Ma W, Wang P, Ma’ayan A. Protocol for using Multiomics2Targets to identify targets and driver kinases for cancer cohorts profiled with multi-omics assays. STAR Protocols 5(4):103457 (2024) PMID: 39565691
Marino GB, Clarke DJB, Lachmann A, Deng EZ, Ma’ayan A. RummaGEO: Automatic mining of human and mouse gene sets from GEO. Patterns (10):101072 (2024) PMID: 39569206
Deng EZ, Marino GB, Clarke DJB, Diamant I, Resnick AC, Ma W, Wang P, Ma’ayan A. Multiomics2Targets identifies targets from cancer cohorts profiled with transcriptomics, proteomics, and phosphoproteomics. Cell Reports Methods 4(8):100839 (2024) PMID: 39127042
Clarke DJB, Evangelista JE, […], Ma’ayan A. Playbook Workflow Builder: interactive construction of bioinformatics workflows from a network of microservices. bioRxiv 2024.06.08.598037 (2024)
Clarke DJB, Marino GB, Deng EZ, Xie Z, Evangelista JE, Ma’ayan A. Rummagene: massive mining of gene sets from supporting materials of biomedical research publications. Communications Biology 7(1):482 (2024) PMID: 38643247
Vinik Y, Maimon A, Dubey V, Raj H, Abramovitch I, Malitsky S, Itkin M, Ma’ayan A, Westermann F, Gottlieb E, Ruppin E, Lev S. Programming a ferroptosis-to-apoptosis transition landscape revealed ferroptosis biomarkers and repressors for cancer therapy. Advanced Science 11(17):e2307263 (2024) PMID: 38441406
Petralia F, Ma W, Yaron TM, Caruso FP, Tignor N, Wang JM, Charytonowicz D, Johnson JL, Huntsman EM, Marino GB, Calinawan A, Evangelista JE, Selvan ME, Chowdhury S, Rykunov D, Krek A, Song X, Turhan B, Christianson KE, Lewis DA, Deng EZ, Clarke DJB, Whiteaker JR, Kennedy JJ, Zhao L, Segura RL, Batra H, Raso MG, Parra ER, Soundararajan R, Tang X, Li Y, Yi X, Satpathy S, Wang Y, Wiznerowicz M, González-Robles TJ, Iavarone A, Gosline SJC, Reva B, Robles AI, Nesvizhskii AI, Mani DR, Gillette MA, Klein RJ, Cieslik M, Zhang B, Paulovich AG, Sebra R, Gümüş ZH, Hostetter G, Fenyö D, Omenn GS, Cantley LC, Ma’ayan A, Lazar AJ, Ceccarelli M, Wang P; Clinical Proteomic Tumor Analysis Consortium. Pan-cancer proteogenomics characterization of tumor immunity. Cell 187(5):1255-1277.e27 (2024) PMID: 38359819
2023
Marino GB, Ahmed N, Xie Z, Jagodnik KM, Han J, Clarke DJB, Lachmann A, Keller MP, Attie AD, Ma’ayan A. D2H2: diabetes data and hypothesis hub. Bioinformatics Advances 3(1):vbad178 (2023) PMID: 38107655
Ma’ayan A, Schlessinger A, Filizola M, Sebra R, Shi Y, Pandey G, Itan Y, Kenny EE. AI + omics are bringing the bench closer to the bedside. Frontiers of Medical Research: Artificial Intelligence (Science/AAAS Washington, DC, 2023) p. 8-10.
Xie Z, Chen C, Ma’ayan A. Dex-Benchmark: datasets and code to evaluate algorithms for transcriptomics data analysis. PeerJ 11:e16351 (2023) PMID: 37953774
Amadori L, Calcagno C, Fernandez DM, Koplev S, Fernandez N, Kaur R, Mury P, Khan NS, Sajja S, Shamailova R, Cyr Y, Jeon M, Hill CA, Chong PS, Naidu S, Sakurai K, Ghotbi AA, Soler R, Eberhardt N, Rahman A, Faries P, Moore KJ, Fayad ZA, Ma’ayan A, Giannarelli C. Systems immunology-based drug repurposing framework to target inflammation in atherosclerosis. Nature Cardiovascular Research 2(6):550-571 (2023) PMID: 37771373
Evangelista JE, Clarke DJB, Xie Z, Marino GB, Utti V, Jenkins SL, Ahooyi TM, Bologa CG, Yang JJ, Binder JL, Kumar P, Lambert CG, Grethe JS, Wenger E, Taylor D, Oprea TI, de Bono B, Ma’ayan A. Toxicology knowledge graph for structural birth defects. Communications Medicine 3(1):98 (2023) PMID: 37460679
Evangelista JE, Xie Z, Marino GB, Nguyen N, Clarke DJB, Ma’ayan A. Enrichr-KG: bridging enrichment analysis across multiple libraries. Nucleic Acids Research 51(W1):W168-W179 (2023) PMID: 37166973
Marino GB, Ngai M, Clarke DJB, Fleishman RH, Deng EZ, Xie Z, Ahmed N, Ma’ayan A. GeneRanger and TargetRanger: processed gene and protein expression levels across cells and tissues for target discovery. Nucleic Acids Research 51(W1):W213-W224 (2023) PMID: 37166966
Deng EZ, Fleishman RH, Xie Z, Marino GB, Clarke DJB, Ma’ayan A. Computational screen to identify potential targets for immunotherapeutic identification and removal of senescence cells. Aging Cell 22(6):e13809 (2023) PMID: 37082798
Marino GB, Wojciechowicz ML, Clarke DJB, Kuleshov MV, Xie Z, Jeon M, Lachmann A, Ma’ayan A. lncHUB2: aggregated and inferred knowledge about human and mouse lncRNAs. Database (Oxford) 2023:baad009 (2023) PMID: 36869839
Lachmann A, Rizzo KA, Bartal A, Jeon M, Clarke DJB, Ma’ayan A. PrismEXP: gene annotation prediction from stratified gene-gene co-expression matrices. PeerJ 11:e14927 (2023) PMID: 36874981
2022
Charbonneau AL, Brady A, Czajkowski K, Aluvathingal J, Canchi S, Carter R, Chard K, Clarke DJB, Crabtree J, Creasy HH, D’Arcy M, Felix V, Giglio M, Gingrich A, Harris RM, Hodges TK, Ifeonu O, Jeon M, Kropiwnicki E, Lim MCW, Liming RL, Lumian J, Mahurkar AA, Mandal M, Munro JB, Nadendla S, Richter R, Romano C, Rocca-Serra P, Schor M, Schuler RE, Tangmunarunkit H, Waldrop A, Williams C, Word K, Sansone SA, Ma’ayan A , Wagner R, Foster I, Kesselman C, Brown CT, White O. Making Common Fund data more findable: catalyzing a data ecosystem. Gigascience 11:giac105 (2022) PMID: 36409836
Clarke DJB, Rebman AW, Fan J, Soloski MJ, Aucott JN, Ma’ayan A. Gene set predictor for post-treatment Lyme disease. Cell Reports Medicine 3(11):100816 (2022) PMID: 36384094
Gross SM, Dane MA, Smith RL, Devlin KL, McLean IC, Derrick DS, Mills CE, Subramanian K, London AB, Torre D, Evangelista JE, Clarke DJB, Xie Z, Erdem C, Lyons N, Natoli T, Pessa S, Lu X, Mullahoo J, Li J, Adam M, Wassie B, Liu M, Kilburn DF, Liby TA, Bucher E, Sanchez-Aguila C, Daily K, Omberg L, Wang Y, Jacobson C, Yapp C, Chung M, Vidovic D, Lu Y, Schurer S, Lee A, Pillai A, Subramanian A, Papanastasiou M, Fraenkel E, Feiler HS, Mills GB, Jaffe JD, Ma’ayan A, Birtwistle MR, Sorger PK, Korkola JE, Gray JW, Heiser LM. A multi-omic analysis of MCF10A cells provides a resource for integrative assessment of ligand-mediated molecular and phenotypic responses. Communications Biology 5(1):1066 (2022) PMID: 36207580
Jenkins EC, Chattopadhyay M, Gomez M, Torre D, Ma’ayan A, Torres-Martin M, Sia D, Germain D. Age alters the oncogenic trajectory toward luminal mammary tumors that activate unfolded proteins responses. Cell Aging 21(10):e13665 (2022) PMID: 36111352
Jeon M, Xie Z, Evangelista JE, Wojciechowicz ML, Clarke DJB, Ma’ayan A. Transforming L1000 profiles to RNA-seq-like profiles with deep learning. BMC Bioinformatics 23(1):74 (2022) PMID: 36100892
Pilarczyk M, Fazel-Najafabadi M, Kouril M, Shamsaei B, Vasiliauskas J, Niu W, Mahi N, Zhang L, Clark NA, Ren Y, White S, Karim R, Xu H, Biesiada J, Bennett MF, Davidson SE, Reichard JF, Roberts K, Stathias V, Koleti A, Vidovic D, Clarke DJB, Schürer SC, Ma’ayan A, Meller J, Medvedovic M. Connecting omics signatures and revealing biological mechanisms with iLINCS. Nature Communications 13(1):4678 (2022) PMID: 35945222
Xie Z, Kropiwnicki E, Wojciechowicz ML, Jagodnik KM, Shu I, Bailey A, Clarke DJB, Jeon M, Evangelista JE, V Kuleshov M, Lachmann A, Parigi AA, Sanchez JM, Jenkins SL, Ma’ayan A. Getting started with LINCS datasets and tools. Current Protocols 2(7):e487 (2022) PMID: 35876555
Evangelista JE, Clarke DJB, Xie Z, Lachmann A, Jeon M, Chen K, Jagodnik KM, Jenkins SL, Kuleshov MV, Wojciechowicz ML, Schurer SC, Medvedovic M, Ma’ayan A. SigCom LINCS: data and metadata search engine for a million gene expression signatures. Nucleic Acids Research 50(W1):W697-709 (2022) PMID: 35524556
Clarke DJB, Kuleshov MV, Xie Z, Evangelista JE, Meyers MR, Kropiwnicki E, Jenkins SL, Ma’ayan A. Gene and drug landing page aggregator. Bioinformatics Advances 2(1):vbac013 (2022) PMID: 35368424
Wickramasinghe NM, Sachs D, Shewale B, Gonzalez DM, Dhanan-Krishnan P, Torre D, LaMarca E, Raimo S, Dariolli R, Serasinghe MN, Mayourian J, Sebra R, Beaumont K, Iyengar S, French DL, Hansen A, Eschenhagen T, Chipuk JE, Sobie EA, Jacobs A, Akbarian S, Ischiropoulos H, Ma’ayan A, Houten SM, Costa K, Dubois NC. PPARdelta activation induces metabolic and contractile maturation of human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 29(4):559-576.e7 (2022) PMID: 35325615
Kropiwnicki E, Lachmann A, Clarke DJB, Xie Z, Jagodnik KM, Ma’ayan A. DrugShot: querying biomedical search terms to retrieve prioritized lists of small molecules. BMC Bioinformatics 23(1):76 (2022) PMID: 35183110
Lachmann A, Xie Z, Ma’ayan A. blitzGSEA: Efficient computation of Gene Set Enrichment Analysis through Gamma distribution approximation. Bioinformatics btac076 (2022) PMID: 35143610
Kropiwnicki E, Binder JL, Yang JJ, Holmes J, Lachmann A, Clarke DJB, Sheils T, Kelleher KJ, Metzger VT, Bologa CG, Oprea TI, Ma’ayan A. Getting started with the IDG KMC datasets and tools. Current Protocols 2(1):e355 (2022) PMID: 35085427
2021
Onoufriadis A, Proudfoot LE, Ainali C, Torre D, Papanikolaou M, Rayinda T, Rashidghamat E, Danarti R, Mellerio JE, Ma’ayan A, McGrath JA. Transcriptomic profiling of recessive dystrophic epidermolysis bullosa wounded skin highlights drug repurposing opportunities to improve wound healing. Experimental Dermatology (2021) PMID: 34694680
Piret SE, Attallah AA, Gu X, Guo Y, Gujarati NA, Henein J, Zollman A, Hato T, Ma’ayan A, Revelo MP, Dickman KG, Chen CH, Shun CT, Rosenquist TA, He JC, Mallipattu SK. Loss of proximal tubular transcription factor Krüppel-like factor 15 exacerbates kidney injury through loss of fatty acid oxidation. Kidney International (2021) PMID: 34634362
Bobe JR, Jutras BL, Horn EJ, Embers ME, Bailey A, Moritz RL, Zhang Y, Soloski MJ, Ostfeld RS, Marconi RT, Aucott J, Ma’ayan A, Keesing F, Lewis K, Ben Mamoun C, Rebman AW, McClune ME, Breitschwerdt EB, Reddy PJ, Maggi R, Yang F, Nemser B, Ozcan A, Garner O, Di Carlo D, Ballard Z, Joung HA, Garcia-Romeu A, Griffiths RR, Baumgarth N, Fallon BA. Recent progress in Lyme disease and remaining challenges. Frontiers in Medicine (2021) PMID: 34485323
Kuleshov MV, Xie Z, London ABK, Yang J, Evangelista JE, Lachmann A, Shu I, Torre D, Ma’ayan A. KEA3: improved kinase enrichment analysis via data integration. Nucleic Acids Research 49(W1):W304-W316 (2021) PMID: 34019655
Piret SE, Guo Y, Attallah AA, Horne SJ, Zollman A, Owusu D, Henein J, Sidorenko VS, Revelo MP, Hato T, Ma’ayan A, He JC, Mallipattu SK. Krüppel-like factor 6-mediated loss of BCAA catabolism contributes to kidney injury in mice and humans. Proc Natl Acad Sci USA 118(23) (2021) PMID: 34074766
Sevilla A, Papatsenko D, Mazloom AR, Xu H, Vasileva A, Unwin RD, LeRoy G, Chen EY, Garrett-Bakelman FE, Lee DF, Trinite B, Webb RL, Wang Z, Su J, Gingold J, Melnick A, Garcia BA, Whetton AD, MacArthur BD, Ma’ayan A, Lemischka IR. An Esrrb and Nanog cell fate regulatory module controlled by feed forward loop interactions. Frontiers in Cell and Developmental Biology 9:630067 (2021) PMID: 33816475
Kropiwnicki E, Evangelista JE, Stein DJ, Clarke DJB, Lachmann A, Kuleshov MV, Jeon M, Jagodnik KM, Ma’ayan A. Drugmonizome and Drugmonizome-ML: integration and abstraction of small molecule attributes for drug enrichment analysis and machine learning. Database baab017 (2021) PMID: 33787872
Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Jeon M, Ma’ayan A. Gene set knowledge discovery with Enrichr. Current Protocols 1(3):e90 (2021) PMID: 33780170
Clarke DJB, Rebman AW, Bailey A, Wojciechowicz ML, Jenkins SL, Evangelista JE, Danieletto M, Fan J, Eshoo MW, Mosel MR, Robinson W, Ramadoss N, Bobe J, Soloski MJ, Aucott JN, Ma’ayan A. Predicting Lyme Disease from patients’ peripheral blood mononuclear cells profiled with RNA-sequencing. Frontiers in Immunology 12:636289 (2021) PMID: 33763080
Austin E, Koo E, Merleev A, Torre D, Marusina A, Luxardi G, Mamalis A, Isseroff RR, Ma’ayan A, Maverakis E, Jagdeo J. Transcriptome analysis of human dermal fibroblasts following red light phototherapy. Scientific Reports 11(1):7315 (2021) PMID: 33795767
Clarke DJB, Jeon M, Stein DJ, Moiseyev N, Kropiwnicki E, Dai C, Xie Z, Wojciechowicz ML, Litz S, Hom J, Evangelista JE, Goldman L, Zhang S, Yoon C, Ahamed T, Bhuiyan S, Cheng M, Karam J, Jagodnik KM, Shu I, Lachmann A, Ayling S, Jenkins SL, Ma’ayan A. Appyters: Turning Jupyter notebooks into data-driven web apps. Patterns 2(3):100213 (2021) PMID: 33748796
Jeon M, Jagodnik KM, Kropiwnicki E, Stein DJ, Ma’ayan A. Prioritizing pain-associated targets with machine learning. Biochemistry (2021) PMID: 33606503
Zhang L, Wang Z, Liu R, Li Z, Lin J, Wojciechowicz ML, Huang J, Lee K, Ma’ayan A, He JC. Connectivity mapping identifies BI-2536 as a potential drug to treat diabetic kidney disease. Diabetes 70(2):589-602 (2021) PMID: 33067313
2020
Eberlein J, Davenport B, Nguyen TT, Victorino F, Jhun K, van der Heide V, Kuleshov M, Ma’ayan A, Kedl R, Homann D. Chemokine signatures of pathogen-specific T cells I: Effector T cells. Journal of Immunology 205(8):2169-2187 (2020) PMID: 32948687
Davenport B, Eberlein J, Nguyen TT, Victorino F, van der Heide V, Kuleshov M, Ma’ayan A, Kedl R, Homann D. Chemokine signatures of pathogen-specific T cells II: Memory T cells in acute and chronic infection. Journal of Immunology 205(8):2188-2206 (2020) PMID: 32948682
Kuleshov MV, Stein DJ, Clarke DJB, Kropiwnicki E, Jagodnik KM, Bartal A, Evangelista JE, Hom J, Cheng M, Bailey A, Zhou A, Ferguson LB, Lachmann A, Ma’ayan A. The COVID-19 gene and drug set library. Patterns 1(6):100090 (2020) PMID: 32838343
Chen Z, Herting CJ, Ross JL, Gabanic B, Vallcorba MP, Szulzewsky F, Wojciechowicz ML, Cimino PJ, Ezhilarasan R, Sulman EP, Ying M, Ma’ayan A, Read RD, Hambardzumyan D. Genetic driver mutations introduced in identical cell-of-origin in murine glioblastoma reveal distinct immune landscapes but similar response to checkpoint blockade. Glia 68(10):2148-2166 (2020) PMID: 32639068
Borziak K, Qi T, Evangelista JE, Clarke DJB, Ma’ayan A, Finkelstein J. Towards intelligent integration and sharing of stem cell research data. Studies in Health Technologies and Informatics 272:334-337 (2020) PMID: 32604670
Wojciechowicz ML, Ma’ayan A. GPR84: an immune response dial? Nature Reviews Drug Discovery 19(6):374 (2020) PMID: 32494048
Bartal A, Lachmann A, Clarke DJB, Seiden AH, Jagodnik KM, Ma’ayan A. EnrichrBot: Twitter bot tracking tweets about human genes. Bioinformatics 36(12):3932-3934 (2020) PMID: 32277816
Bernitz JM, Rapp K, Daniel MG, Shcherbinin D, Yuan Y, Gomes A, Waghray A, Brosh R, Lachmann A, Ma’ayan A, Papatsenko D, Moore KA. Memory of divisional history directs the continuous process of primitive hematopoietic lineage commitment. Stem Cell Reports 14(4):561-574 (2020) PMID: 32243840
Lachmann A, Clarke DJB, Torre D, Xie Z, Ma’ayan A. Interoperable RNA-seq analysis in the cloud. Biochimica et Biophysica Acta Gene Regulatory Mechanisms 1863(6):194521 (2020) PMID: 32156561
Edwards JJ, Rouillard AD, Fernandez NF, Wang Z, Lachmann A, Shankaran SS, Bisgrove BW, Demarest B, Turan N, Srivastava D, Bernstein D, Deanfield J, Giardini A, Porter G, Kim R, Roberts AE, Newburger JW, Goldmuntz E, Brueckner M, Lifton RP, Seidman CE, Chung WK, Tristani-Firouzi M, Yost HJ, Ma’ayan A, Gelb BD. Systems analysis implicates WAVE2 complex in the pathogenesis of developmental left-sided obstructive heart defects. JACC Basic to Translational Science 5(4):376-386 (2020) PMID: 32368696
Heitman N, Sennett R, Mok KW, Saxena N, Srivastava D, Martino P, Grisanti L, Wang Z, Ma’ayan A, Rompolas P, Rendl M. Dermal sheath contraction powers stem cell niche relocation during hair cycle regression. Science 367(6474):161-166 (2020) PMID: 31857493
2019
Clarke DJB, Wang L, Jones A, Wojciechowicz ML, Torre D, Jagodnik KM, Jenkins SL, McQuilton P, Flamholz z, […], Ma’ayan A. FAIRshake: Toolkit to evaluate the FAIRness of research digital resources. Cell Systems 9(5):417-421 (2019) PMID: 31677972
Ochsner SA, Abraham D, Martin K, Ding W, McOwiti A, Kankanamge W, Wang Z, Andreano K, Hamilton RA, Chen Y, Hamilton A, Gantner ML, Dehart M, Qu S, Hilsenbeck SG, Becnel LB, Bridges D, Ma’ayan A, Huss JM, Stossi F, Foulds CE, Kralli A, McDonnell DP, McKenna NJ. The Signaling Pathways Project, an integrated ‘omics knowledgebase for mammalian cellular signaling pathways. Scientific Data 6(1):252 (2019) PMID: 31672983
Duncan A, Heyer MP, Ishikawa M, Caligiuri SPB, Liu XA, Chen Z, Vittoria Micioni Di Bonaventura M, Elayouby KS, Ables JL, Howe WM, Bali P, Fillinger C, Williams M, O’Connor RM, Wang Z, Lu Q, Kamenecka TM, Ma’ayan A, O’Neill HC, Ibanez-Tallon I, Geurts AM, Kenny PJ. Habenular TCF7L2 links nicotine addiction to diabetes. Nature 574(7778):372-377 (2019) PMID: 31619789
Fernandez DM, Rahman AH, Fernandez NF, Chudnovskiy A, Amir ED, Amadori L, Khan NS, Wong CK, Shamailova R, Hill CA, Wang Z, Remark R, Li JR, Pina C, Faries C, Awad AJ, Moss N, Bjorkegren JLM, Kim-Schulze S, Gnjatic S, Ma’ayan A, Mocco J, Faries P, Merad M, Giannarelli C. Single-cell immune landscape of human atherosclerotic plaques. Nature Medicine (10):1576-1588 (2019) PMID: 31591603
Niepel M, Hafner M, Mills CE, Subramanian K, Williams EH, Chung M, Gaudio B, Barrette AM, Stern AD, Hu B, Korkola JE, LINCS Consortium et al. A Multi-center study on the reproducibility of drug-response assays in mammalian cell lines. Cell Systems 9(1):35-48.e5 (2019) PMID: 31302153
Lachmann A, Schilder BM, Wojciechowicz ML, Torre D, Kuleshov MV, Keenan AB, Ma’ayan A. Geneshot: search engine for ranking genes from arbitrary text queries. Nucleic Acids Research 47(W1):W571-W577 (2019) PMID: 31114885
Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz ML, Utti V, Jagodnik KM, Kropiwnicki E, Wang Z, Ma’ayan A. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Research 47(W1):W212-W224 (2019) PMID: 31114921
Kuleshov MV, Diaz JEL, Flamholz ZN, Keenan AB, Lachmann A, Wojciechowicz ML, Cagan RL, Ma’ayan A. modEnrichr: a suite of gene set enrichment analysis tools for model organisms. Nucleic Acids Research 47(W1):W183-W190 (2019) PMID: 31069376
Nakahara F, Borger DK, Wei Q, Pinho S, Maryanovich M, Zahalka AH, Suzuki M, Cruz CD, Wang Z, Xu C, Boulais PE, Ma’ayan A, Greally JM, Frenette PS. Engineering a haematopoietic stem cell niche by revitalizing mesenchymal stromal cells. Nature Cell Biology 21(5):560-567 (2019) PMID: 30988422
Keenan AB, Wojciechowicz ML, Wang Z, Jagodnik KM, Jenkins SL, Lachmann A, Ma’ayan A. Connectivity mapping: methods and applications. Annual Review of Biomedical Data Science 2:69-92 (2019)
Tohme R, Izadmehr S, Gandhe S, Tabaro G, Vallabhaneni S, Thomas A, Vasireddi N, Dhawan NS, Ma’ayan A, Sharma N, Galsky MD, Ohlmeyer M, Sangodkar J, Narla G. Direct activation of PP2A for the treatment of tyrosine kinase inhibitor-resistant lung adenocarcinoma. JCI Insight 4(4) pii: 125693 (2019) PMID: 30830869
Ellis RJ, Wang Z, Genes N, Ma’ayan A. Predicting opioid dependence from electronic health records with machine learning. BioData Mining 12:3 (2019) PMID: 30728857
Mok KW, Saxena N, Heitman N, Grisanti L, Srivastava D, Muraro MJ, Jacob T, Sennett R, Wang Z, Su Y, Yang LM, Ma’ayan A, Ornitz DM, Kasper M, Rendl M. Dermal condensate niche fate specification occurs prior to formation and is placode progenitor dependent. Developmental Cell 48(1):32-48 (2019) PMID: 30595537
Wang Z, Lachmann A, Ma’ayan A. Mining data and metadata from the gene expression omnibus. Biophysical Reviews 11(1):103-110 (2019) PMID: 30594974
2018
Torre D, Lachmann A, Ma’ayan A. BioJupies: Automated Generation of Interactive Notebooks for RNA-Seq Data Analysis in the Cloud. Cell Systems 7(5):556-561 (2018) PMID: 30447998
Oprea TI, Jan L, Johnson GL, Roth BL, Ma’ayan A, Schurer S, Shoichet BK, Sklar LA, McManus MT. Far away from the lamppost. PLoS Biology 16(12):e3000067 (2018) PMID: 30532236
Breen MS, Ozcan S, Ramsey JM, Wang Z, Ma’ayan A, Rustogi N, Gottschalk MG, Webster MJ, Weickert CS, Buxbaum JD, Bahn S. Temporal proteomic profiling of postnatal human cortical development. Translational Psychiatry 8(1):267 (2018) PMID: 30518843
Gomes AM, Kurochkin I, Chang B, Daniel M, Law K, Satija N, Lachmann A, Wang Z, Ferreira L, Ma’ayan A, Chen BK, Papatsenko D, Lemischka IR, Moore KA, Pereira CF. Cooperative Transcription Factor Induction Mediates Hemogenic Reprogramming. Cell Reports 25(10):2821-2835 (2018) PMID: 30517869
Wang Z, He E, Sani K, Jagodnik KM, Silverstein M, Ma’ayan A. Drug Gene Budger (DGB): An application for ranking drugs to modulate a specific gene based on transcriptomic signatures. Bioinformatics (2018) PMID: 30169739
Strub T, Ghiraldini FG, Carcamo S, Li M, Wroblewska A, Singh R, Goldberg MS, Hasson D, Wang Z, Gallagher SJ, Hersey P, Ma’ayan A, Long GV, Scolyer RA, Brown B, Zheng B, Bernstein E. SIRT6 haploinsufficiency induces BRAFV600E melanoma cell resistance to MAPK inhibitors via IGF signalling. Nature Communications 9(1):3440 (2018) PMID: 30143629
Guo Y, Pace J, Li Z, Ma’ayan A, Wang Z, Revelo MP, Chen E, Gu X, Attalah A, Yang Y, Estrada C, Yang VW, He JC, Mallipattu SK. Podocyte-specific induction of Krüppel-Like Factor 15 restores differentiation markers and attenuates kidney injury in proteinuric kidney disease.. Journal of the America Society of Nephrology pii: ASN.2018030324(2018) PMID: 30143559
Clarke DJB, Kuleshov MV, Schilder BM, Torre D, Duffy ME, Keenan AB, Lachmann A, Feldmann AS, Gundersen GW, Silverstein MC, Wang Z, Ma’ayan A. eXpression2Kinases (X2K) Web: linking expression signatures to upstream cell signaling networks. Nucleic Acids Research 46(W1):W171-W179 (2018) PMID: 29800326
Han S, Tan C, Ding J, Wang J, Ma’ayan A, Gouon-Evans V. Endothelial cells instruct liver specification of embryonic stem cell-derived endoderm through endothelial VEGFR2 signaling and endoderm epigenetic modifications. Stem Cell Research 30:163-170 (2018) PMID: 29936335
Stathias V, Koleti A, Vidović D, Cooper DJ, Jagodnik KM, Terryn R, Forlin M, Chung C, Torre D, Ayad N, Medvedovic M, Ma’ayan A, Pillai A, Schurer SC. Sustainable data and metadata management at the BD2K-LINCS Data Coordination and Integration Center. Scientific Data 5:180117 (2018) PMID: 29917015
Grimes M, Hall B, Foltz L, Levy T, Rikova K, Gaiser J, Cook W, Smirnova E, Wheeler T, Clark NR, Lachmann A, Zhang B, Hornbeck P, Ma’ayan A, Comb M. Integration of protein phosphorylation, acetylation, and methylation data sets to outline lung cancer signaling networks. Science Signaling 11(531) (2018) PMID: 29789295
Lachmann A, Torre D, Keenan AB, Jagodnik KM, Lee HJ, Wang L, Silverstein MC, Ma’ayan A. Massivc mining of publicly available RNA-sq data from human and mouse. Nature Communications 9(1):1366 (2018) PMID: 29636450
Torre D, Krawczuk P, Jagodnik KM, Lachmann A, Wang Z, Wang L, Kuleshov MV, Ma’ayan A. Datasets2Tools, repository and search engine for bioinformatics datasets, tools and canned analyses. Scientific Data 5, 180023 (2018) PMID: 29485625
Oprea TI, Bologa CG, Brunak S, Campbell A, Gan GN, Gaulton A, Gomez SM, Guha R, Hersey A, Holmes J, Jadhav A, Jensen LJ, Johnson GL, Karlson A, Leach AR, Ma’ayan A et al. Unexplored therapeutic opportunities in the human genome. Nature Review Drug Discovery (2018) PMID: 29572638
Czarnowicki T, Dohlman AB, Malik K, Antonini D, Bissonnette R, Chan TC, Zhou L, Wen HC, Estrada Y, Xu H, Bryson C, Shen J, Lala D, Ma’ayan A, McGeehan G, Gregg R, Guttman-Yassky E. Short-term LXR activation improves epidermal barrier features in mild-to-moderate atopic dermatitis: a randomized controlled trial. Annals of Allergy, Asthma and Immunology S1081-1206(18)30211-4 (2018) PMID: 29567358
Wang Z, Lachmann A, Keenan AB, Ma’ayan A. L1000FWD: Fireworks visualization of drug-induced transcriptomic signatures. Bioinformatics 34(12)2150-2152 (2018) PMID: 29420694
Koplev S, Lin K, Dohlman AB, Ma’ayan A. Integration of pan-cancer transcriptomics with RPPA proteomics reveals mechanisms of epithelial-mesenchymal transition. PLoS Computational Biology 14(1):e1005911 (2018) PMID: 29293502
Hodos R, Zhang P, Lee HC, Duan Q, Wang Z, Clark NR, Ma’ayan A, Wang F, Kidd B, Hu J, Sontag D, Dudley J. Cell-specific prediction and application of drug-induced gene expression profiles. Pacific Symposium on Biocomputing 23:32-43 (2018) PMID: 29218867
Keenan AB, Jenkins SL, Jagodnik KM, Koplev S, He E, Torre D, Wang Z, Dohlman AB, Silverstein MC, Lachmann A, Kuleshov MV, Ma’ayan A et al. The Library of Integrated Network-Based Cellular Signatures NIH program: System-level cataloging of human cells response to perturbations. Cell Systems 6(1):13-24 (2018) PMID: 29199020
Koleti A, Terryn R, Stathias V, Chung C, Cooper DJ, Turner JP, Vidovic D, Forlin M, Kelley TT, D’Urso A, Allen BK, Torre D, Jagodnik KM, Wang L, Jenkins SL, Mader C, Niu W, Fazel M, Mahi N, Pilarczyk M, Clark N, Shamsaei B, Meller J, Vasiliauskas J, Reichard J, Medvedovic M, Ma’ayan A, Pillai A, Schurer SC. Data Portal for the Library of Integrated Network-based Cellular Signatures (LINCS) program: integrated access to diverse large-scale cellular perturbation response data. Nucleic Acids Research 46(D1):D558-D566 (2018) PMID: 29140462
2017
Wang Z, Li L, Glicksberg BS, Israel A, Dudley JT, Ma’ayan A. Predicting age by mining electronic medical records with deep learning characterizes differences between chronological and physiological age. Journal of Biomedical Informatics pii: S1532-0464(17)30240-X (2017) PMID: 29113935
Fu J, Wang Z, Lee K, Wei C, Liu Z, Zhang M, Zhoud M, Cai M, Zhang W, Chuang PY, Ma’ayan A, He JC, Liu Z. Transcriptomic analysis uncovers novel synergistic mechanisms in combination therapy for lupus nephritis. Kidney International pii: S0085-2538(17)30671-3 (2017) PMID: 29102373
Niepel M, Hafner M, Duan Q, Wang Z, Paull EO, Chung M, Lu X, Stuart JM, Golub TR, Subramanian A, Ma’ayan A, Sorger PK. Common and cell-type specific responses to anti-cancer drugs revealed by high throughput transcript profiling. Nature Communications 8(1):1186 (2017) PMID: 29084964
Fernandez NF, Gundersen GW, Rahman A, Grimes ML, Rikova K, Hornbeck P, Ma’ayan A. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Scientific Data 4:170151 (2017) PMID: 28994825
Ma’ayan A. Complex systems biology. Journal of the Royal Society, Interface 14(134) pii: 20170391 (2017) PMID: 28931638
Lachmann A, Torre D, Keenan AB, Jagodnik KM, Lee HJ, Silverstein MC, Wang L, Ma’ayan A. Massive mining of publicly available RNA-seq data from human and mouse. bioRxiv 189092 (2017)
Sangodkar J, Perl A, Tohme R, Kiselar J, Kastrinsky DB, Zaware N, Izadmehr S, Mazhar S, Wiredja DD, O’Connor CM, Hoon D, Dhawan NS, Schlatzer D, Yao S, Leonard D, Borczuk AC, Gokulrangan G, Wang L, Svenson E, Farrington CC, Yuan E, Avelar RA, Stachnik A, Smith B, Gidwani V, Giannini HM, McQuaid D, McClinch K, Wang Z, Levine AC, Sears RC, Chen EY, Duan Q, Datt M, Haider S, Ma’ayan A, DiFeo A, Sharma N, Galsky MD, Brautigan DL, Ioannou YA, Xu W, Chance MR, Ohlmeyer M, Narla G. Activation of tumor suppressor protein PP2A inhibits KRAS-driven tumor growth. The Journal of Clinical Investigation pii: 89548 (2017) PMID: 28504649
Jagodnik KM, Koplev S, Jenkins S, Ohno-Machado L, Paten B, Schurer SC, Dumontier M, Verborgh R, Bui A, Ping P, McKenna NJ, Madduri R, Pillai A, Ma’ayan A. Developing a Framework for Digital Objects in the Big Data to Knowledge (BD2K) Commons: Report from the Commons Framework Pilots Workshop. Journal of Biomedical Informatics pii: S1532-0464(17)30101-6 (2017) PMID: 28501646
Asada N, Kunisaki Y, Pierce H, Wang Z, Fernandez NF, Birbrair A, Ma’ayan A, Frenette PS. Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nature Cell Biology 19(3):214-22 (2017) PMID: 28218906
Shameer K, Glicksberg BS, Hodos R, Johnson KW, Badgeley MA, Readhead B, Tomlinson MS, O’Connor T, Miotto R, Kidd BA, Chen R, Ma’ayan A, Dudley JT. Systematic analyses of drugs and disease indications in RepurposeDB reveal pharmacological, biological and epidemiological factors influencing drug repositioning. Briefings in Bioinformatics (2017) PMID: 28200013
Nguyen DT, Mathias S, Bologa C, Brunak S, Fernandez N, Gaulton A, Hersey A, Holmes J, Jensen LJ, Karlsson A, Liu G, Ma’ayan A, Mandava G, Mani S, Mehta S, Overington J, Patel J, Rouillard AD, Schürer S, Sheils T, Simeonov A, Sklar LA, Southall N, Ursu O, Vidovic D, Waller A, Yang J, Jadhav A, Oprea TI, Guha R. Pharos: Collating protein information to shed light on the druggable genome. Nucleic Acids Research 45(D1):D995-D100 (2017) PMID: 27903890
2016
Pfau ML, Purushothaman I, Feng J, Golden SA, Aleyasin H, Lorsch ZS, Cates HM, Flanigan ME, Menard C, Heshmati M, Wang Z, Ma’ayan A, Shen L, Hodes GE, Russo SJ. Integrative Analysis of Sex-Specific microRNA Networks Following Stress in Mouse Nucleus Accumbens. Frontiers in Molecular Neuroscience 9:144 (2016) PMID: 28066174
Gundersen GW, Jagodnik KM, Woodland H, Fernandez NF, Sani K, Dohlman AB, Ung PM, Monteiro CD, Schlessinger A, Ma’ayan A. GEN3VA: aggregation and analysis of gene expression signatures from related studies. BMC Bioinformatics 17(1):461 (2016) PMID: 27846806
Wang Z, Monteiro CD, Jagodnik KM, Fernandez NF, Gundersen GW, Rouillard AD, Jenkins SL, Feldmann AS, Hu KS, McDermott MG, Duan Q, Clark NR, Jones MR, Kou Y, Goff T, Woodland H, Amaral FM, Szeto GL, Fuchs O, Schüssler-Fiorenza Rose SM, Sharma S, Schwartz U, Bausela XB, Szymkiewicz M, Maroulis V, Salykin A, Barra CM, Kruth CD, Bongio NJ, Mathur V, Todoric RD, Rubin UE, Malatras A, Fulp CT, Galindo JA, Motiejunaite R, Jüschke C, Dishuck PC, Lahl K, Jafari M, Aibar S, Zaravinos A, Steenhuizen LH, Allison LR, Gamallo P, de Andres Segura F, Dae Devlin T, Pérez-García V, Ma’ayan A. Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd. Nature Communications 7:12846 (2016) PMID: 27667448
Duan Q, Reid SP, Clark NR, Wang Z, Fernandez NF, Rouillard AD, Readhead B, Tritsch SR, Hodos R, Hafner M, Niepel M, Sorger PK, Dudley JT, Bavari S, Panchal RG, Ma’ayan A. L1000CDS2: LINCS L1000 characteristic direction signatures search engine. npj Systems Biology and Applications 2, 16015 (2016)
von Schimmelmann M, Feinberg PA, Sullivan JM, Ku SM, Badimon A, Duff MK, Wang Z, Lachmann A, Dewell S, Ma’ayan A, Han MH, Tarakhovsky A, Schaefer A. Polycomb repressive complex 2 (PRC2) silences genes responsible for neurodegeneration. Nature Neuroscience 19(10):1321-30 (2016) PMID: 27526204
Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, Ma’ayan A. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) pii: baw100 (2016) PMID: 27374120
Wang Z, Ma’ayan A. An open RNA-Seq data analysis pipeline tutorial with an example of reprocessing data from a recent Zika virus study. F1000Research 5:1574 (2016) PMID: 27583132
Mallipattu SK, Guo Y, Revelo MP, Roa-Peña L, Miller T, Ling J, Shankland SJ, Bialkowska AB, Ly V, Estrada C, Jain MK, Lu Y, Ma’ayan A, Mehrotra A, Yacoub R, Nord EP, Woroniecki RP, Yang VW, He JC. Krüppel-Like Factor 15 Mediates Glucocorticoid-Induced Restoration of Podocyte Differentiation Markers. Journal of the American Society of Nephrology pii: ASN.2015060672 (2016) PMID: 27288011
Morgan DJ, Poolman TM, Williamson AJ, Wang Z, Clark NR, Ma’ayan A, Whetton AD, Brass A, Matthews LC, Ray DW. Glucocorticoid receptor isoforms direct distinct mitochondrial programs to regulate ATP production. Scientific Reports 6:26419 (2016) PMID: 27226058
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott M, Monteiro CD, Gundersen GW, Ma’ayan A. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Research 44(W1):W90-7 (2016) PMID: 27141961
Wang Z, Clark NR, Ma’ayan A. Drug Induced Adverse Events Prediction with the LINCS L1000 Data. Bioinformatics 32(15):2338-45 (2016) PMID: 27153606
Rezza A, Wang Z, Sennett R, Qiao W, Wang D, Heitman N, Mok KW, Clavel C, Yi R, Zandstra P, Ma’ayan A, Rendl M. Signaling Networks among Stem Cell Precursors, Transit-Amplifying Progenitors, and their Niche in Developing Hair Follicles. Cell Reports pii: S2211-1247(16)30213-3 (2016) PMID: 27009580
Scheckel C, Drapeau E, Frias MA, Park CY, Fak J, Zucker-Scharff I, Kou Y, Haroutunian V, Ma’ayan A, Buxbaum JD, Darnell RB. Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain. Elife pii: e10421 (2016) PMID: 26894958
Kong DS, Kim J, Lee IH, Kim ST, Seol HJ, Lee JI, Park WY, Ryu G, Wang Z, Ma’ayan A, Nam DH. Integrative radiogenomic analysis for multicentric radiophenotype in glioblastoma. Oncotarget 7(10):11526-38 (2016) PMID: 26863628
Khan JA, Mendelson A, Kunisaki Y, Birbrair A, Kou Y, Arnal-Estapé A, Pinho S, Ciero P, Nakahara F, Ma’ayan A, Bergman A, Merad M, Frenette PS. Fetal liver hematopoietic stem cell niches associate with portal vessels. Science 351(6269):176-80 (2016) PMID: 26634440
2015
Clark NR, Szymkiewicz M, Wang Z, Monteiro CD, Jones MR, Ma’ayan A. Principal Angle Enrichment Analysis (PAEA): Dimensionally reduced multivariate gene set enrichment analysis tool. Proceedings (IEEE Int Conf Bioinformatics Biomed) (2015) PMID: 26848405
Liang R, Campreciós G, Kou Y, McGrath K, Nowak R, Catherman S, Bigarella CL, Rimmele P, Zhang X, Gnanapragasam MN, Bieker JJ, Papatsenko D, Ma’ayan A, Bresnick E, Fowler V, Palis J, Ghaffari S. A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal Erythropoiesis. PLoS Genetics 11(10):e1005526 (2015) PMID: 26452208
Sennett R, Wang Z, Rezza A, Grisanti L, Roitershtein N, Sicchio C, Mok KW, Heitman NJ, Clavel C, Ma’ayan A, Rendl M. An Integrated Transcriptome Atlas of Embryonic Hair Follicle Progenitors, Their Niche, and the Developing Skin. Developmental Cell 34(5):577-91 (2015) PMID:26256211
Stockton SD Jr, Gomes I, Liu T, Moraje C, Hipólito L, Jones MR, Ma’ayan A, Morón JA, Li H, Devi LA. Morphine regulated synaptic networks revealed by integrated proteomics and network analysis. Molecular and Cellular Proteomics pii: mcp.M115.047977 (2015) PMID: 26149443
Rouillard AD, Wang Z, Ma’ayan A. Abstraction for data integration: Fusing mammalian molecular, cellular and phenotype big datasets for better knowledge extraction. Computational Biology and Chemistry pii: S1476-9271(15)00083-3 (2015) PMID: 26297300
Wang Z, Clark NR, Ma’ayan A. Dynamics of the discovery process of protein-protein interactions from low content studies. BMC Systems Biology 9(1):26 (2015) PMID: 26048415
Gundersen GW, Jones MR, Rouillard AD, Kou Y, Monteiro CD, Feldmann AS, Hu KS, Ma’ayan A. GEO2Enrichr: browser extension and server app to extract gene sets from GEO and analyze them for biological functions. Bioinformatics pii: btv297 (2015) PMID: 25971742
Mallipattu SK, Horne SJ, D’Agati V, Narla G, Liu R, Frohman MA, Dickman K, Chen EY, Ma’ayan A, Bialkowska AB, Ghaleb AM, Nandan MO, Jain MK, Daehn I, Chuang PY, Yang VW, He JC. Kruppel-like factor 6 regulates mitochondrial function in the kidney. Journal of Clinical Investigation 215(3):1347-61 (2015) PMID: 25689250
2014
Duarte LF, Young AR, Wang Z, Wu HA, Panda T, Kou Y, Kapoor A, Hasson D, Mills NR, Ma’ayan A, Narita M, Bernstein E. Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nature Communications 5:5210 (2014) PMID: 25394905
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Ercument Cicek A, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Fu SC, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Crooks L, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Sean Hill R, Ionita-Laza I, Jimenez Gonzalez P, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei J, Lehtimaki T, Lin CF, Ma’ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515(7526):209-15 (2014) PMID: 25363760
Ma’ayan A, Duan Q. A blueprint of cell identity. Nature Biotechnology 32(10):1007-1008 (2014) PMID: 25299921
Xu H, Ang YS, Sevilla A, Lemischka IR, Ma’ayan A. Construction and validation of a regulatory network for pluripotency and self-renewal of mouse embryonic stem cells. PLoS Computational Biology 10(8):e1003777 (2014) PMID: 25122140
Ma’ayan A, Rouillard AD, Clark NR, Wang Z, Duan Q, Kou Y. Lean Big Data integration in systems biology and systems pharmacology. Trends in Pharmacological Sciences 35(9):450-460 (2014) PMID: 25109570
Duan Q, Wang Z, Fernandez NF, Rouillard AD, Tan CM, Benes CH, Ma’ayan A. Drug/Cell-line Browser (DCB): Interactive Canvas Visualization of Cancer Drug/Cell-Line Viability Assay Datasets. Bioinformatics pii: btu526 (2014) PMID: 25100688
Hays T, Ma’ayan A, Clark NR, Tan CM, Teixeira A, Teixeira A, Choi JW, Burdis N, Jung SY, Bajaj AO, O’Malley BW, He JC, Hyink DP, Klotman PE. Proteomics Analysis of the Non-Muscle Myosin Heavy Chain IIa-Enriched Actin-Myosin Complex Reveals Multiple Functions within the Podocyte. PLoS One 9(6):e100660 (2014) PMID: 24949636
Duan Q, Flynn C, Niepel M, Hafner M, Muhlich JL, Fernandez NF, Rouillard AD, Tan CM, Chen EY, Golub TR, Sorger PK, Subramanian A, Ma’ayan A. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Research pii: gku476 (2014) PMID: 24906883
Clark NR, Hu KS, Feldmann AS, Kou Y, Chen EY, Duan Q, Ma’ayan A. The characteristic direction: a geometrical approach to identify differentially expressed genes. BMC Bioinformatics 15:79 (2014) PMID: 24650281
Liu L, Lei J, Sanders SJ, Willsey AJ, Kou Y, Cicek AE, Klei L, Lu C, He X, Li M, Muhle RA, Ma’ayan A, Noonan JP, Sestan N, McFadden KA, State MW, Buxbaum JD, Devlin B, Roeder K. DAWN: A framework to identify autism genes and subnetworks using gene expression and genetics. Molecular Autism 5(1):22 (2014) PMID: 24602502
Baker SJ, Ma’ayan A, Lieu YK, John P, Reddy MV, Chen EY, Duan Q, Snoeck HW, Reddy EP. B-myb is an essential regulator of hematopoietic stem cell and myeloid progenitor cell development. Proc Natl Acad Sci U S A 111(8):3122-7 (2014) PMID: 24516162
Azeloglu EU, Hardy SV, Eungdamrong NJ, Chen Y, Jayaraman G, Chuang PY, Fang W, Xiong H, Neves SR, Jain MR, Li H, Ma’ayan A, Gordon RE, He JC, Iyengar R. Interconnected network motifs control podocyte morphology and kidney function. Science Signaling 7(311):ra12 (2014) PMID: 24497609
Kleensang A, Maertens A, Rosenberg M, Fitzpatrick S, Lamb J, Auerbach S, Brennan R, Crofton KM, Gordon B, Fornace Jr AJ, Gaido K, Gerhold D, Haw R, Henney A, Ma’ayan A, McBride M, Monti S, Ochs MF, Pandey A, Sharan R, Stierum R, Tugendreich S, Willett C, Wittwehr C, Xia J, Patton GW, Arvidson K, Bouhifd M, Hogberg HT, Luechtefeld T, Smirnova L, Zhao L, Adeleye Y, Kanehisa M, Carmichael P, Andersen ME, Hartung T. t4 Workshop Report: Pathways of Toxicity. ALTEX 31(1):53-61 (2014) PMID: 24127042
2013
Poultney CS, Goldberg AP, Drapeau E, Kou Y, Harony-Nicolas H, Kajiwara Y, De Rubeis S, Durand S, Stevens C, Rehnström K, Palotie A, Daly MJ, Ma’ayan A, Fromer M, Buxbaum JD. Identification of Small Exonic CNV from Whole-Exome Sequence Data and Application to Autism Spectrum Disorder. American Journal of Human Genetics 93(4):607-619 (2013) PMID: 24094742
Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, Ivanov S, Duan Q, Bala S, Condon T, van Rooijen N, Grainger JR, Belkaid Y, Ma’ayan A, Riches DW, Yokoyama WM, Ginhoux F, Henson PM, Randolph GJ. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity 39(3):599-610 (2013) PMID: 24012416
Xu H, Baroukh C, Dannenfelser R, Chen EY, Tan CM, Kou Y, Kim YE, Lemischka IR, Ma’ayan A. ESCAPE: database for integrating high-content published data collected from human and mouse embryonic stem cells. Database (Oxford) bat045 (2013) PMID: 23794736
Pereira CF, Chang B, Qiu J, Niu X, Papatsenko D, Hendry CE, Clark NR, Nomura-Kitabayashi A, Kovacic JC, Ma’ayan A, Schaniel C, Lemischka IR, Moore K. Induction of a Hemogenic Program in Mouse Fibroblasts. Cell Stem Cell 13(2):205-18 (2013) PMID: 23770078
Tan CM, Chen EY, Dannenfelser R, Clark NR, Ma’ayan A. Network2Canvas: network visualization on a canvas with enrichment analysis. Bioinformatics 29(15):1872-8 (2013) PMID: 23749960
Sillivan SE, Whittard JD, Jacobs MM, Ren Y, Mazloom AR, Caputi FF, Horvath M, Keller E, Ma’ayan A, Pan YX, Chiang LW, Hurd YL. ELK1 Transcription Factor Linked to Dysregulated Striatal Mu Opioid Receptor Signaling Network and OPRM1 Polymorphism in Human Heroin Abusers. Biological Psychiatry 74(7):511-9 (2013) PMID: 23702428
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14:128 (2013) PMID: 23586463
Zhong Y, Chen EY, Liu R, Chuang PY, Mallipattu SK, Tan CM, Clark NR, Deng Y, Klotman PE, Ma’ayan A, He JC. Renoprotective Effect of Combined Inhibition of Angiotensin-Converting Enzyme and Histone Deacetylase. Journal of the American Society of Nephrology 24(5):801-11(2013) PMID: 23559582
Duan Q, Kou Y, Clark NR, Gordonov S, Ma’ayan A. Metasignatures identify two major subtypes of breast cancer. CPT: Pharmacometrics & Systems Pharmacology 2:e35 (2013) PMID: 23836026
Jenkins SL, Ma’ayan A. Systems pharmacology meets predictive, preventive, personalized and participatory medicine. Pharmacogenomics 14(2):119-22 (2013) PMID: 23327571
Mallipattu SK, Liu R, Zhong Y, Chen EY, D’Agati V, Kaufman L, Ma’ayan A, Klotman PE, Chuang PY, He JC. Expression of HIV transgene aggravates kidney injury in diabetic mice. Kidney International 83(4):626-34 (2013) PMID: 23325078
2012
Ma’ayan A. Colliding dynamical complex network models: Biological attractors versus attractors from material physics. Biophysical Journal 103(9):1816-1817 (2012) PMID: 23199907
Rabinowitz KM, Wang Y, Chen E, Hovhannisyan Z, Chiang D, Berin MC, Dahan S, Chaussabel D, Ma’ayan A, Mayer L. Transforming Growth Factor-b Signaling Controls Activities of Human Intestinal CD8 T Suppressor Cells. Gastroenterology pii: S0016-5085(12)01744-1 (2012) PMID: 23232296
Clavel C, Grisanti L, Zemla R, Rezza A, Barros R, Sennett R, Mazloom AR, Chung CY, Cai X, Cai CL, Pevny L, Nicolis S, Ma’ayan A, Rendl M. Sox2 in the Dermal Papilla Niche Controls Hair Growth by Fine-Tuning BMP Signaling in Differentiating Hair Shaft Progenitors. Developmental Cell 23(5):981-94 (2012) PMID: 23153495
Macarthur BD, Sevilla A, Lenz M, Müller FJ, Schuldt BM, Schuppert AA, Ridden SJ, Stumpf PS, Fidalgo M, Ma’ayan A, Wang J, Lemischka IR. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nature Cell Biology 14(11):1139-47 (2012) PMID: 23103910
Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma’ayan A, Chua WJ, Hansen TH, Turley SJ, Merad M, Randolph GJ; the Immunological Genome Consortium. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature Immunology 13(11):1118-1128 (2012) PMID: 23023392
Clark NR, Dannenfelser R, Tan CM, Komosinski ME, Ma’ayan A. Sets2Networks: network inference from repeated observations of sets. BMC Systems Biology 6(1):89 (2012) PMID 22824380
Zhou Q, Lv H, Mazloom AR, Xu H, Ma’ayan A, Gallo JM. Activation of Alternate Pro-Survival Pathways Accounts for Acquired Sunitinib Resistance in U87MG Glioma Xenografts. Journal of Pharmacology and Experimental Therapeutics 343(2):509-19 (2012) PMID: 22869928
Lee DF, Su J, Ang YS, Carvajal-Vergara X, Mulero-Navarro S, Pereira CF, Gingold J, Wang HL, Zhao R, Sevilla A, Darr H, Williamson AJ, Chang B, Niu X, Aguilo F, Flores ER, Sher YP, Hung MC, Whetton AD, Gelb BD, Moore KA, Snoeck HW, Ma’ayan A, Schaniel C, Lemischka IR. Regulation of Embryonic and Induced Pluripotency by Aurora Kinase-p53 Signaling. Cell Stem Cell 11(2):179-94 (2012) PMID: 22862944
Dannenfelser R, Clark NR, Ma’ayan A. Genes2FANs: connecting genes through functional association networks. BMC Bioinformatics 2;13(1):156 (2012) PMID: 22748121
Kou Y, Betancur C, Xu H, Buxbaum JD, Ma’ayan A. Network- and attribute-based classifiers can prioritize genes and pathways for autism spectrum disorders and intellectual disability. American Journal of Medical Genetics Part C: Seminars in Medical Genetics 160C(2):130-42 (2012) PMID: 22499558
Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485(7397):242-5 (2012) PMID: 22495311
Mallipattu SK, Liu R, Zheng F, Narla G, Ma’ayan A, Dikman S, Jain MK, Saleem M, D’Agati V, Klotman P, Chuang PY, He JC. Kruppel-Like factor 15 (KLF15) is a key regulator of podocyte differentiation. Journal of Biological Chemistry 287(23):19122-35 (2012) PMID: 22493483
Jin Y, Ratnam K, Chuang PY, Fan Y, Zhong Y, Dai Y, Mazloom AR, Chen EY, D’Agati V, Xiong H, Ross MJ, Chen N, Ma’ayan A, He JC. A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nature Medicine 18(4):580-8 (2012) PMID: 22406746
Rozenfeld R, Bushlin I, Gomes I, Tzavaras N, Gupta A, Neves S, Battini L, Gusella GL, Lachmann A, Ma’ayan A, Blitzer RD, Devi LA. Receptor heteromerization expands the repertoire of cannabinoid signaling in rodent neurons. PLoS One 7(1):e29239 (2012) PMID: 22235275
Chen EY, Xu H, Gordonov S, Lim MP, Perkins MH, Ma’ayan A. Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics 28(1):105-11 (2012) PMID: 22080467
He JC, Chuang PY, Ma’ayan A, Iyengar R. Systems biology of kidney diseases. Kidney International 81(1):22-39 (2012) PMID: 21881558
Ding J, Xu H, Faiola F, Ma’ayan A, Wang J. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Research 22(1):155-67 (2012) PMID: 22083510
2011
Mazloom AR, Dannenfelser R, Clark NR, Grigoryan AV, Linder KM, Cardozo TJ, Bond JC, Boran ADW, Iyengar R, Malovannaya A, Lanz RB, Ma’ayan A. Recovering protein-protein and domain-domain interactions from aggregation of IP-MS proteomics of coregulator complexes. PLoS Computational Biology 7(12):e1002319 (2011) PMID: 22219718
Xu H, Ma’ayan A. Visualization of Patient Samples by Dimensionality Reduction of Genome-Wide Measurements. Lecture Notes Computer Science 7058:15-22 (2011)
Abul-Husn NS, Annangudi SP, Ma’ayan A, Ramos-Ortolaza DL, Stockton SD Jr, Gomes I, Sweedler JV, Devi LA. Chronic morphine alters the presynaptic protein profile: identification of novel molecular targets using proteomics and network analysis. PLoS One 6(10):e25535 (2011) PMID: 22043286
Baroukh C, Jenkins SL, Dannenfelser R, Ma’ayan A. Genes2WordCloud: a quick way to identify biological themes from gene lists and free text. Source Code for Biology and Medicine 6(1):15 (2011) PMID: 21995939
Ma’ayan A. Introduction to network analysis in systems biology. Science Signaling 4(190):tr5 (2011) PMID: 21917719
Clark NR, Ma’ayan A. Introduction to statistical methods for analyzing large data sets: gene-set enrichment analysis. Science Signaling 4(190):tr4 (2011) PMID: 21917718
Clark NR, Ma’ayan A. Introduction to statistical methods to analyze large data sets: principal components analysis. Science Signaling 4(190):tr3 (2011) PMID: 21917717
Dannenfelser R, Lachmann A, Szenk M, Ma’ayan A. FNV: Light-weight Flash-based network and pathway viewer. Bioinformatics 27(8):1181-2 (2011) PMID: 21349871
Webb RL, Ma’ayan A. Sig2BioPAX: Java tool for converting flat files to BioPAX Level 3 format. Source Code for Biology and Medicine 6:5 (2011) PMID: 21418653
2010
Xu H, Lemischka IR, Ma’ayan A. SVM classifier to predict genes important for self-renewal and pluripotency of mouse embryonic stem cells. BMC Systems Biology 4(1):173 (2010) PMID: 21176149
Carcea I, Ma’ayan A, Mesias R, Sepulveda B, Salton SR, Benson DL. Flotillin-mediated endocytic events dictate cell type-specific responses to Semaphorin 3A. Journal of Neuroscience 30(45):15317-29 (2010) PMID: 21068336
Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26(19):2438-44 (2010) PMID: 20709693
Méndez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466(7308):829-34 (2010) PMID: 20703299
Xiao K, Sun J, Kim J, Rajagopal S, Zhai B, Villén J, Haas W, Kovacs JJ, Shukla AK, Hara MR, Hernandez M, Lachmann A, Zhao S, Lin Y, Cheng Y, Mizuno K, Ma’ayan A, Gygi SP, Lefkowitz RJ. Global phosphorylation analysis of {beta}-arrestin-mediated signaling downstream of a seven transmembrane receptor (7TMR). Proc Natl Acad Sci U S A 107(34):15299-304 (2010) PMID: 20686112
Ma’ayan A, He JC. Protein kinase target discovery from genome-wide messenger RNA expression profiling. Mount Sinai Journal of Medicine 77(4):345-9 (2010) PMID: 20687179
MacArthur BD, Sanchez-Garcia RJ, Ma’ayan A. Microdynamics and criticality of adaptive regulatory networks. Physical Review Letters 104(16):168701 (2010) PMID: 20482087
Berger SI, Ma’ayan A, Iyengar R. Systems pharmacology of arrhythmias. Science Signaling 3(118):ra30 (2010) PMID:20407125
Hernandez M, Lachmann A, Zhao S, Xiao K, Ma’ayan A. Inferring the sign of kinase-substrate interactions by combining SILAC phosphoproteomics with a literature-based mammalian kinome network. 10th IEEE International Conference on Bioinformatics and BioEngineering (2010) BIBE.
Xu H, Schaniel C, Lemischka IR, Ma’ayan A. Toward a complete in-silico, multi-layered embryonic stem cell regulatory network. Wiley Interdisciplinary Reviews: Systems Biology and Medicine 2(6):708-33 (2010) PMID 20890967
Lachmann A, Ma’ayan A. Lists2Networks: Integrated analysis of gene/protein lists. BMC Bioinformatics 11:87 (2010) PMID: 20152038
MacArthur BD, Lachmann A, Lemischka IR, Ma’ayan A. GATE: Software for the analysis and visualization of high-dimensional time-series expression data. Bioinformatics 26(1) 143-144 (2010) PMID: 19892805
2009
Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma’ayan A, Boyer LA, Troyanskaya OG, Whetton AD, Lemischka IR. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature 462(7271) 358-362 (2009) PMID: 19924215
Cordeddu V, Di Schiavi E, Pennacchio LA, Ma’ayan A, Sarkozy A, Fodale V, Cecchetti S, Cardinale A, et al. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nature Genetics. 41(9) 1022-6 (2009) PMID: 19684605
MacArthur BD, Ma’ayan A, Lemischka IR. Systems biology of stem cell fate and cellular reprogramming. Nature Reviews: Molecular Cell Biology (10) 672-81 (2009) PMID: 19738627
Abul-Husn NS, Bushlin I, Morón JA, Jenkins SL, Dolios G, Wang R, Iyengar R, Ma’ayan A, Devi LA. Systems approach to explore components and interactions in the presynapse. Proteomics 9(12):3303-15 (2009) PMID: 19562802
Lachmann A, Ma’ayan A. KEA: Kinase enrichment analysis. Bioinformatics 25(5) 684-6 (2009) PMID: 19176546
Ma’ayan A, Jenkins SL, Barash A, Iyengar R. Neuro2A differentiation by Galphai/o pathway. Science Signaling 2(54):cm1 (2009) PMID: 19155528
Ma’ayan A, Jenkins SL, Webb RL, Berger SI, Purushothaman SP, Abul-Husn N, Posner JM, Flores T, Iyengar R. SNAVI: Desktop Application for Analysis and Visualization of Large-Scale Signaling Networks. BMC Systems Biology 3;10 (2009) PMID: 19154595
Ma’ayan A. Insights into the organization of biochemical regulatory networks using graph theory analyses. Journal of Biological Chemistry 284(9) 5451-5 (2009) PMID: 18940806
2008
Ma’ayan A, Cecchi GA, Wagner J, Rao AR, Iyengar R, Stolovitzky G. Ordered Cyclic Motifs Contributes to Dynamic Stability in Biological and Engineered Networks. Proc Natl Acad Sci U S A 105(49) 19235-40 (2008) PMID: 19033453
MacArthur BD, Ma’ayan A, Lemischka IR. Toward stem cell systems biology: from molecules to networks and landscapes. Cold Spring Harbor Symposium on Quantitative Biology 73;211-5 (2008) PMID: 19329576
Ma’ayan A. Network integration and graph analysis in mammalian molecular systems biology. IET Systems Biology 2(5) 206-21 (2008) PMID: 19045817
Lu TC, Wang Z, Feng X, Chuang P, Fang W, Chen Y, Neves S, Ma’ayan A, Xiong H, Liu Y, Iyengar R, Klotman PE, He JC. Retinoic acid utilizes CREB and USF-1 in a transcriptional feed-forward loop in order to stimulate MKP1 expression in HIV-infected podocytes. Molecular and Cellular Biology 28(18) 5785-94 (2008) PMID: 18625721
Bromberg KD, Ma’ayan A, Neves SR, Iyengar R. Design of a CB1 receptor signaling network to trigger neurite outgrowth. Science 320, 903-909 (2008) PMID: 18487186
Ma’ayan A, Lipshtat A, Iyengar R, Sontag ED. Proximity of Intracellular Regulatory Networks to Monotone Systems. IET Systems Biology 2, 103-112 (2008) PMID: 18537452
Ma’ayan A. PubMedAlertMe – Standalone Windows-based PubMed SDI Software. Computers in Biology and Medicine 38:620-622 (2008) PMID: 18402930
Lipshtat A, Purushothaman SP, Iyengar R, Ma’ayan A. Functions of bifans in context of multiple regulatory motifs in signaling network. Biophysical Journal 94, 2566-2579 (2008) PMID: 18178648
2007
White AG, Ma’ayan A. Connecting seed lists of mammalian proteins using steiner trees. ACSSC 2007. Conference Record of the Forty-First Asilomar Conference on Signals, Systems and Computers, 2007. 4-7 Nov. 2007 page(s): 155-159 Pacific Grove, CA, USA
Caplan AJ, Ma’ayan A, Willis IM. Multiple kinases and system robustness: A link between Cdc37 and genome integrity. Cell Cycle 6:3145 (2007) PMID: 18075309
Berger SI, Posner JM, Ma’ayan A. Genes2Networks: connecting lists of proteins by using background literature-based mammalian networks. BMC Bioinformatics 8:372 (2007) PMID: 17916244
Berger SI, Iyengar R, Ma’ayan A. AVIS: AJAX Viewer of Interactive Signaling Networks. Bioinformatics 23, 2803-5 (2007) PMID: 17855420
Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional Atlas of the Integrin Adhesome. Nature Cell Biology 9, 858 (2007) PMID: 17671451
Ma’ayan A, Jenkins SL, Goldfarb J, Iyengar R. Network analysis of FDA approved drugs and their targets. Mount Sinai Journal of Medicine 74, 27 (2007) PMID: 17516560
Hasseldine A, Lipshtat A, Iyengar R, Ma’ayan A. (2007) Modeling Cell Signaling Networks. In “Bioinformatics – From Genomes to Therapies” Vol. 2 Edited by Thomas Lengauer. Pages 829- 860 (WILEY-VCH, Weinheim, Germany)
2006
Ma’ayan A, Gardiner K, Iyengar R. The cognitive phenotype of Down syndrome: Insights from intracellular network analysis. NeuroRx 3, 396 (2006) PMID: 16815222
Ma’ayan A, Lipshtat A, Iyengar R. Topology of resultant signaling network shaped by evolutionary pressure. Physical Review: E, Statistical, Nonlinear and Soft Matter Physics 73, 061912 (2006) PMID: 16906869
Ma’ayan A, Iyengar R. From components to regulatory network motifs in signaling networks. Briefings in Functional Genomics and Proteomics 5, 57 (2006) PMID: 16769680
2005
Ma’ayan A, Blitzer RD, Iyengar R. Toward predictive models of mammalian cells. Annual Review of Biophysics and Biomolecular Structure 34, 319 (2005) PMID: 15869393
Ma’ayan A, Jenkins SL, Neves S, Hasseldine A, Grace E, Dubin-Thaler B, Eungdamrong NJ, Weng G, Ram PT, Rice JJ, Kershenbaum A, Stolovitzky GA, Blitzer RD, Iyengar R. Formation of regulatory patterns during signal propagation in a Mammalian cellular network. Science 309 (5737)1078 (2005) PMID: 16099987

