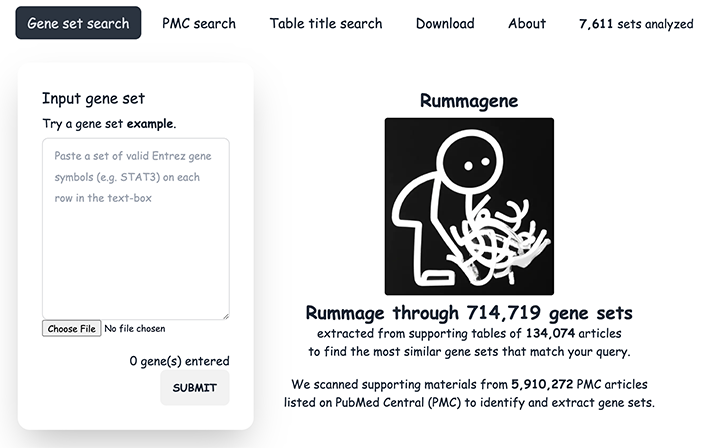
The Ma’ayan Laboratory conducts multi-disciplinary NIH funded research that utilizes Big Data analytics to develop better understanding about drug action in human cells, build molecular regulatory networks from high-content genome-wide data, and predict optimized therapeutics for individual patients across several complex diseases.
Bioinformatics Software Engineer
Posted November 2025
A full-time position as a Bioinformatics Software Engineer is available in the Ma’ayan Laboratory of Computational Systems Biology and the Mount Sinai Center for Bioinformatics at the Icahn School of Medicine at Mount Sinai in New York.
What you’ll do:
The successful candidate will collaborate with an interdisciplinary team on developing, implementing, documenting and maintaining web-based software applications used by the larger scientific community. As a member of our team, we would like you to be able to:
-
- Work independently to identify and define technical requirements for tasks and timelines
- Design, build, test, and deploy scalable bioinformatics web-based applications in a cloud environment
- Develop, document, and maintain version-controlled code
- Mock, develop, and enhance interactive UI designs
- Build Docker containers for various bioinformatics workflows
- Maintain and enhance efficient solutions to reproducible workflow orchestrations on the cloud and local HPC
- Author and manage technical documentation that concisely describes design and implementation details
- Manage, publish and maintain code repository (eg. GitHub), container repository (eg. DockerHub)
- Respond to new feature requests, assist with issues raised by userbase as needed
- Report project status regularly to the Principal Investigator
What you’ll bring:
-
- Bachelor’s or Master’s degree in Computer Science, Informatics, Mathematics, Statistics, Engineering or Biomedical Science
- Knowledge of open-source bioinformatics tools and workflows
- Experience working with high performance clusters and cloud technologies
- Experience developing web-based applications with front ends utilizing frameworks such as React, NodeJS, RShiny, Flask, or Dash
- Experience with building and orchestrating containers (Docker) using technologies like Kubernetes
- Extensive experience with Git or other version control systems
- Experience in more than one programming language such as Python, JavaScript, Java, C/C++, R
- Working knowledge of relational and non-relational databases
- Strong communication (written and verbal) and organizational skills
To apply, please e-mail your CV/resume and the contact information of three references to: sherry.jenkins@mssm.edu
Postdoctoral Fellow, Big Data Science and Computational Systems Biology
Posted November 2025
A full-time postdoctoral position is available in the Ma’ayan Laboratory of Computational Systems Biology and the Mount Sinai Center for Bioinformatics at the Icahn School of Medicine at Mount Sinai in New York.
What you’ll do:
The successful candidate will collaborate with an interdisciplinary team to develop tools and algorithms for the analysis, integration, and visualization of large scale biological omics datasets. The datasets include genomics, transcriptomics, epigenomics, proteomics, and metabolomics. In addition, the position involves the application of machine learning, including deep learning, to mining electronic medical records and combining such data with omics datasets.
What you’ll bring:
Candidates are required to have a recent PhD in Biomedical Science, Computer Science, Mathematics, Biostatistics, Statistics, Physics, Engineering, and relevant experience with applications to biology.
-
- Experience with machine learning, multithread programming, and cloud computing
- Experience developing and deploying web-based and mobile apps
- Experience with bioinformatics research projects
- Knowledge of Python, R, Java, JavaScript, Node.js, MongoDB, MySQL, Docker
To apply, please e-mail your CV, research statement, and contact information of three references to: sherry.jenkins@mssm.edu
Bioinformatician II
Posted November 2025
A full-time position is available in the Ma’ayan Laboratory of Computational Systems Biology and the Mount Sinai Center for Bioinformatics at the Icahn School of Medicine at Mount Sinai in New York.
What you’ll do:
The successful candidate will collaborate with an interdisciplinary team on projects related to bioinformatics, big data science, and systems biology including developing, implementing, documenting and maintaining web-based software applications used by the larger scientific community. You will work on various aspects of research and infrastructure projects. Your work will include:
- Developing novel dynamic data visualizations
- Applying machine learning to identify patterns in large and complex datasets
- Harmonizing and abstracting data from a variety of sources
- Developing novel statistical mining strategies and algorithms
- Developing websites, databases, APIs and other data exchange protocols
What you’ll bring:
- Master’s degree in Computer Science, Informatics, Mathematics, Statistics, Physics, Engineering or Biological Sciences and a strong interest in working on data-intensive biomedical problems.
- Experience with machine learning, multithread programming, and cloud computing
- Experience developing and deploying web-based and mobile apps
- Experience with bioinformatics research projects
- Knowledge of Python, R, Java, JavaScript, Node.js, MongoDB, MySQL, Docker
- Knowledge of molecular and cell biology
To apply, please e-mail your CV, research statement, and contact information of three references to: sherry.jenkins@mssm.edu
Bioinformatician I
Posted November 2025
A full-time position is available in the Ma’ayan Laboratory of Computational Systems Biology and the Mount Sinai Center for Bioinformatics at the Icahn School of Medicine at Mount Sinai in New York.
What you’ll do:
The successful candidate will collaborate with an interdisciplinary team on projects related to bioinformatics, big data science, and systems biology including developing, implementing, documenting and maintaining web-based software applications used by the larger scientific community. You will work on various aspects of research and infrastructure projects. Your work will include:
- Developing novel dynamic data visualizations
- Applying machine learning to identify patterns in large and complex datasets
- Harmonizing and abstracting data from a variety of sources
- Developing novel statistical mining strategies and algorithms
- Developing websites, databases, APIs and other data exchange protocols
What you’ll bring:
- Bachelor’s degree in Computer Science, Informatics, Mathematics, Statistics, Physics, Engineering, or Biological Sciences and a strong interest in working on data-intensive biomedical problems.
- Experience with machine learning, multithread programming, and cloud computing
- Experience developing and deploying web-based and mobile apps
- Experience with bioinformatics research projects
- Knowledge of Python, R, Java, JavaScript, Node.js, MongoDB, MySQL, Docker
- Knowledge of molecular and cell biology
To apply, please e-mail your CV, research statement, and contact information of three references to: sherry.jenkins@mssm.edu
Current Graduate Students
Posted July 2025
If you are interested in joining the lab as a graduate student, please email Dr. Ma’ayan at avi.maayan@mssm.edu. The Ma’ayan Laboratory accepts rotation students from all Multidisciplinary Training Areas (MTAs) within the ISMMS Graduate School of Biomedical Sciences.
Prospective Graduate Students
Posted July 2025
Prospective graduate students should apply to one of the programs at the ISMMS Graduate School of Biomedical Sciences.