Computational and Mathematical Methods to Study the Complexity of Regulatory Networks in Mammalian Cells
The Ma’ayan Laboratory applies machine learning and other statistical mining techniques to study how intracellular regulatory systems function as networks to control cellular processes such as differentiation, dedifferentiation, apoptosis and proliferation.
Our research team develops software systems to help experimental biologists form novel hypotheses from high-throughput data, while aiming to better understand the structure and function of regulatory networks in mammalian cellular and multi-cellular systems. Read More
Featured Publication
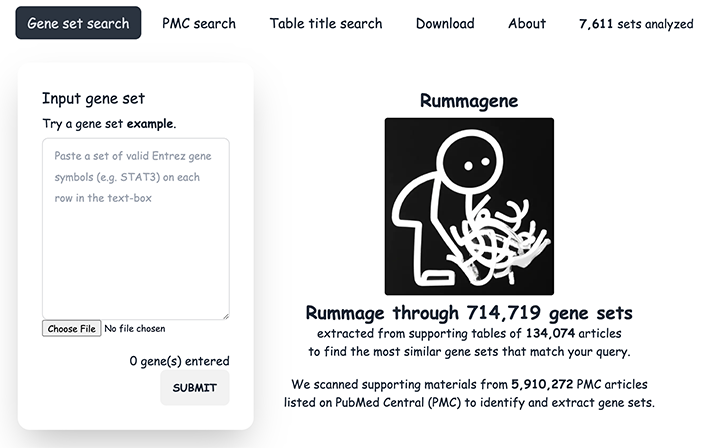
Rummagene: massive mining of gene sets from supporting materials of biomedical research publications
Many biomedical research publications contain gene sets in their supporting tables, and these sets are currently not available for search and reuse. By crawling PubMed Central, the Rummagene server provides access to hundreds of thousands of such mammalian gene sets. These sets are served for enrichment analysis, free text, and table title search. Investigating statistical patterns within the Rummagene database, we demonstrate that Rummagene can be used for transcription factor and kinase enrichment analyses, and for gene function predictions. By combining gene set similarity with abstract similarity, Rummagene can find surprising relationships between biological processes, concepts, and named entities. Overall, Rummagene brings to surface the ability to search a massive collection of published biomedical datasets that are currently buried and inaccessible. The Rummagene web application is available at rummagene.com
Citation: Clarke DJB, Marino GB, Deng EZ, Xie Z, Evangelista JE, Ma’ayan A. Rummagene: massive mining of gene sets from supporting materials of biomedical research publications. Communications Biology 2024 7(1):482.
