Research
Projects: Current & Future
Whole Mount Development
The spatial properties of neural structures—long, thin, and complexly oriented in three-dimensional space—pose unique challenges to imaging. Capturing these structures in their entirety requires high spatial resolution imaging through large depths and fields of view. Restricted by these limitations, traditional approaches slice organs into two-dimensional cross-sections to examine their interior.
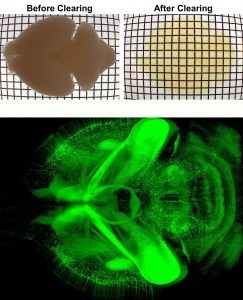 Over the past seven years, we have developed novel whole mount imaging techniques to see inside organs without cutting or deforming their three-dimensional structure. Pairing tissue clearing—the process of making a tissue transparent—with immunohistochemistry, we label proteins of interest inside intact tissue to understand the identity, distribution, and spatial arrangement of both neuronal and non-neuronal cells. We image these cleared tissues using lightsheet microscopy. Our foundational method, iDISCO, was developed in 2014 for imaging intact embryos and brains. We have since continued to further develop our method, releasing iDISCO+ in 2016, and to optimize our approach for application to various peripheral organs, including adipose tissues which are traditionally difficult to clear due to their high lipid content (see AdipoClear, 2018).
Over the past seven years, we have developed novel whole mount imaging techniques to see inside organs without cutting or deforming their three-dimensional structure. Pairing tissue clearing—the process of making a tissue transparent—with immunohistochemistry, we label proteins of interest inside intact tissue to understand the identity, distribution, and spatial arrangement of both neuronal and non-neuronal cells. We image these cleared tissues using lightsheet microscopy. Our foundational method, iDISCO, was developed in 2014 for imaging intact embryos and brains. We have since continued to further develop our method, releasing iDISCO+ in 2016, and to optimize our approach for application to various peripheral organs, including adipose tissues which are traditionally difficult to clear due to their high lipid content (see AdipoClear, 2018).
We apply this technique to study a range of biological questions, both in our lab and with collaborators. The iDISCO family of approaches is not only useful for addressing questions about extensive, complex structures, such as neurons or vasculature, but also offers distinct advantages when studying disease processes where the location of one’s target is not known a priori by facilitating an unbiased survey of the bulk tissue.
3D Proteomics
The mammalian brain contains numerous diverse cell types and functional states, which may undergo changes during disease processes. We are developing a three-dimensional proteomic database of the mouse brain captured at cellular resolution to provide fundamental knowledge of cell type distributions and structural morphology in the healthy adult mouse. These proteomic profiles will link new knowledge generated by single cell transcriptomic profiling to functional analyses. To build this reference, we have established an efficient IHC pipeline to screen monoclonal antibodies (mAbs) for compatibility with whole-mount labeling and imaging based on our advanced iDISCO methods. We are building our reference using monoclonal antibodies because of the advantages these reagents provide in consistent quality and reproducibility. Employing these antibodies, we generate 2D and 3D datasets through whole mount imaging.
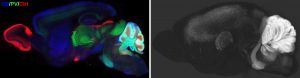
Our purpose in this project is three-fold: 1) To provide 3D whole mount proteomic datasets which can directly be used by other scientists to facilitate versatile molecular analyses of specific brain structures. 2) To build a reliable mAb reference list for numerous protein targets, to contribute to knowledge of reliable reagents and reproducibility within the field. 3) To disseminate optimized protocols with reference IHC results to direct the successful utilization of each mAb clone.
By sharing our resulting proteomic datasets on our publicly-accessible online platform (coming soon), we hope our reference can provide a valuable resource for the field.
Axon Guidance in Neurodevelopment
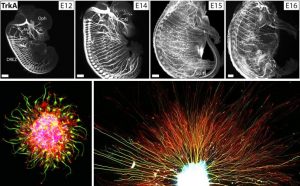
We study how neural axon guidance establishes properly-formed neural circuitry in the developing mouse embryo. We use both in vivo genetics and in vitro culture models of embryonic neurons to investigate the mechanisms by which extending axons are able to navigate precisely in the complex environment and reach their proper targets over long distances. We are interested in understanding how myriad environmental cues (canonical guidance molecules and other ECM factors) orchestrate the dynamic axonal responses to form diverse neural projections. Studying this process will not only help us to understand the blueprint of adult neural circuitry formation to ensure mature functions, but also provide critical insights for mechanisms underlying later axonal plasticity, pruning and degeneration during aging and disease.
Selected Publications
Reconstruction of 1,000 projection neurons reveals new cell types and organization of long-range connectivity in the mouse brain.
Winnubst J, Bas E, Ferreira TA, Wu Z, Economo MN, Edson P, Arthur BJ, Bruns C, Rokicki K, Schauder D, Olbris DJ, Murphy SD, Ackerman DG, Arshadi C, Baldwin P, Blake R, Elsayed A, Hasan M, Ramirez D, Dos Santos B, Weldon M, Zafar A, Dudmann JT, Gerfen CR, Hantman AW, Korff W, Sternson SM, Spruston N, Svoboda K, Chandrashekar J.
Cell (2019) 179:268-281. doi:10.1016/j.cell.2019.07.042 [PMID: 31495573] [PDF]
Long-range guidance of spinal commissural axons by Netrin1 and Sonic hedgehog from midline floor plate cells.
Wu Z, Makihara S, Yam PT, Teo S, Renier N, Balekoglu N, Moreno-Bravo JA, Olsen O, Chédotal A, Charron F, Tessier-Lavigne M.
Neuron (2019) 101:635-647. doi:10.1016/j.neuron.2018.12.025. [PMID: 30661738] [Full Article]
Adipo-Clear: A Tissue Clearing Method for Three-Dimensional Imaging of Adipose Tissue.
Chi J, Crane A, Wu Z, Cohen P. (2018)
Vis. Exp. (2018) (137), e58271, doi:10.3791/58271.
Three-dimensional adipose tissue imaging reveals regional variation in beige fat biogenesis and PRDM16-dependent sympathetic neurite density.
Chi J, Wu Z, Choi C, Nguyen L, Tegegne S, Ackerman SE, Crane A, Marchildon F, Tessier-Lavigne M, and Cohen P†.
Cell Metab. (2018) 27:226-236. doi:10.1016/j.cmet.2017.12.011. [PMID: 29320703] [Full Article]
Mapping of brain activity by automated volume analysis of immediate early genes.
Renier N, Adams EL, Kirst C, Wu Z, Azevedo R, Kohl J, Autry AE, Kadiri L, Venkataraju KU, Zhou Y, Wang VX, Tang CY, Olsen O, Dulac C, Osten P, and Tessier-Lavigne M.
Cell (2016) 165:1789-802. doi:10.1016/j.cell.2016.05.007. [PMID: 27238021] [Full Article]
iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging.
Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M.
Cell (2014) 159:896-910. doi:10.1016/j.cell.2014.10.010. [PMID: 25417164] [Full Article]
Laboratory of Neural Systems, Structures and Genetics
Zhuhao Wu, PhD
zhuhao.wu@mssm.edu