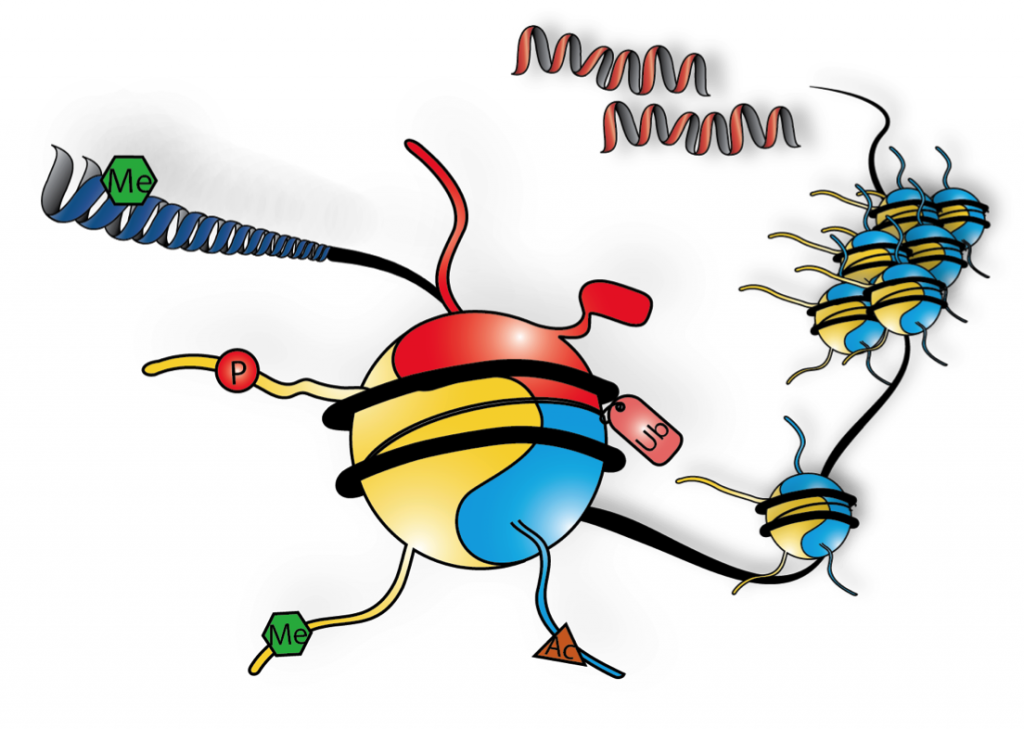
Chromatin is the complex of DNA and its intimately associated proteins -with histones constituting the major component. This template is an attractive candidate for shaping the features of a cell’s epigenetic landscape. Disruption of a cell’s epigenetic balance can perturb chromatin structure and gene regulation, contributing to disease states.
Our mechanistic work focuses on the study of 1) histone variants and 2) ATP-dependent chromatin remodeling complexes in cancer.
1) When incorporated into chromatin, histone variants participate in diverse nuclear functions including centromeric regulation, DNA damage responses, and transcriptional control. Histone variants alter the structure and stability of the nucleosome and provide the cell with the potential to change its post-translational modification (PTM) profile due to amino acid sequence differences from their conventional histone counterparts. We study variants of the H2A and H3 families and their dedicated chaperones that escort these proteins in and out of the chromatin template in healthy cells and in a variety of cancers.
2) Our work also investigates the role of ATP-dependent chromatin remodeling complexes, which are altered at high frequencies in certain cancers. Such complexes perturb the histone-DNA interaction, allowing nucleosome mobilization and chromatin accessibility for proteins, such as transcription factors. We are studying the cellular consequences of mutations and structural alterations in chromatin remodeling factors in melanoma and neuroblastoma.