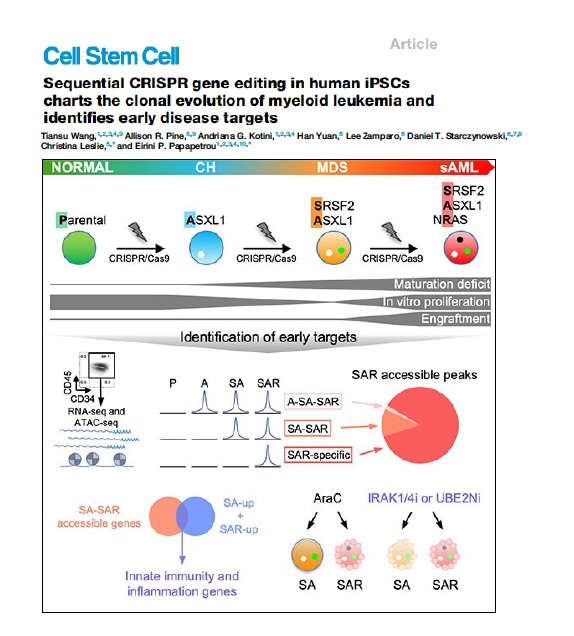
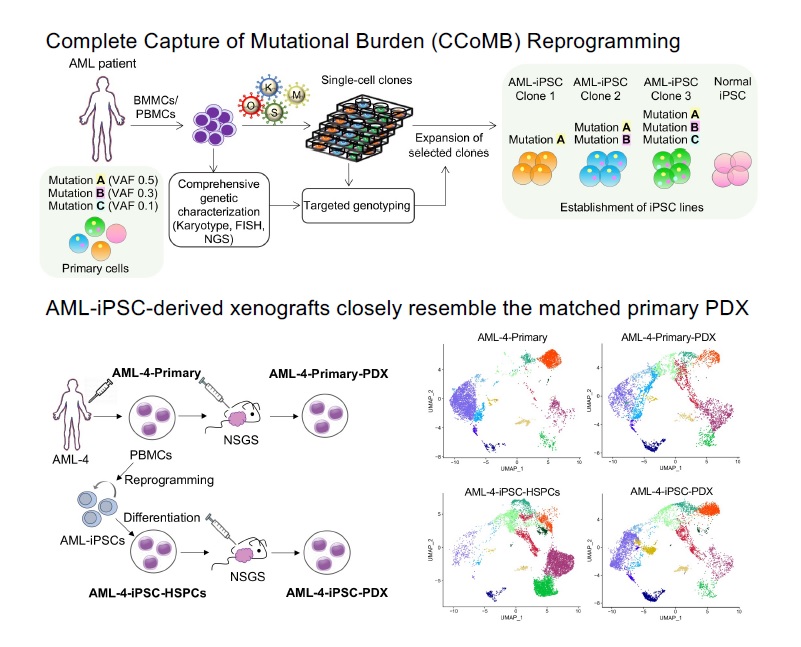
Clonal evolution of AML
AML develops through the step-wise acquisition of driver mutations by a hematopoietic stem or progenitor cell (HSPC), which clonally expands while gradually gaining malignant features. We have created unique iPSC-based models to study this clonal evolution process, using (1) reprogramming of patient cells into iPSCs, and (2) CRISPR/Cas9-mediated precise gene editing to introduce or correct driver mutations in isogenic iPSCs. The leukemia cells derived from these iPSCs closely resemble primary patient cells and enable us to perform functional in vitro studies, as well as in vivo xenograft studies to discover new disease biology and new targets for therapeutic development. We have more recently developed temporally controlled and reversible mutant alleles to address two fundamental questions: (1) How does the order of mutational acquisition affect leukemogenesis? (2) Are fully transformed AML cells still dependent on preleukemic mutations?
Read More
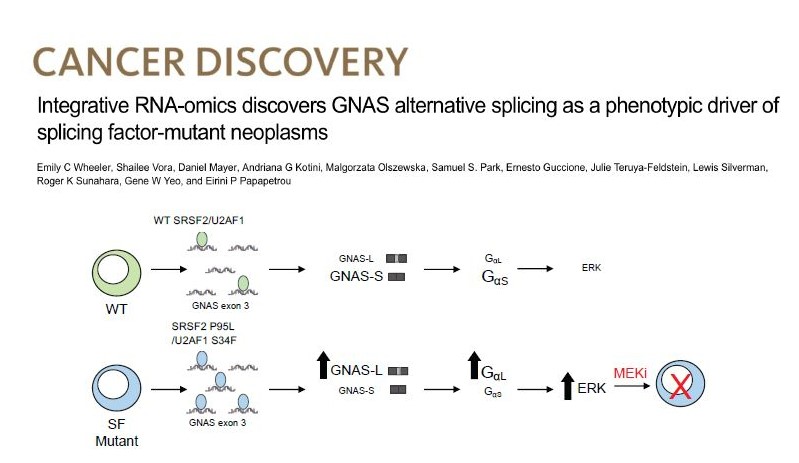
Splicing factor mutations in MDS
Splicing factor (SF) mutations are the most common and disease-defining class of somatic mutations in MDS, but how they drive disease is not understood. We created isogenic human iPSC models of SF mutations with allele-specific epitope tags and performed integrative analyses of mRNA binding and altered splicing. We discovered that SRSF2 and U2AF1 mutations converge in promoting a long isoform of the gene GNAS, which encodes a longer and more active form of the G protein subunit Gαs and activates MAPK/ERK signaling. We are currently pursuing various ways to therapeutically target GNAS/Gαs.
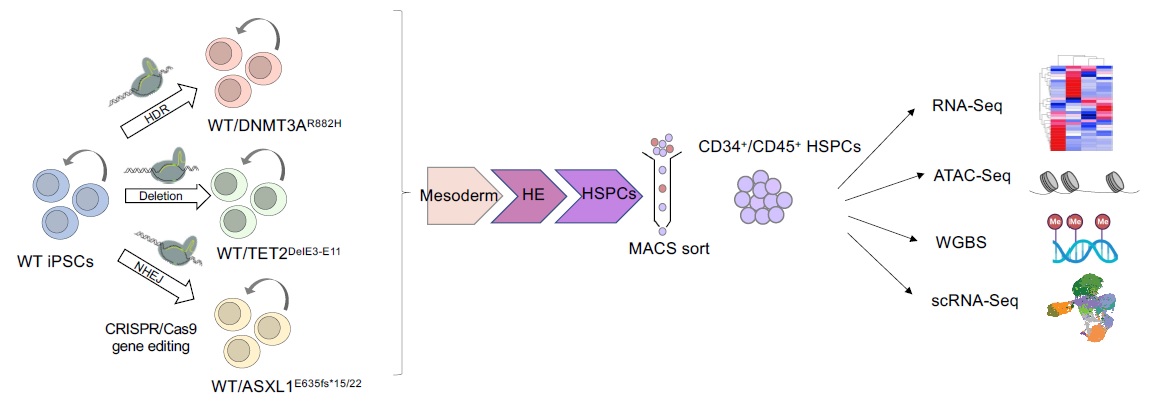
Clonal hematopoiesis
We have generated isogenic iPSC models of the 3 main CH mutations – TET2, DNMT3A and ASXL1 – and are performing integrative multi-omics analyses to identify cell-autonomous effects that underlie the clonal advantage of HSPCs that carry them.