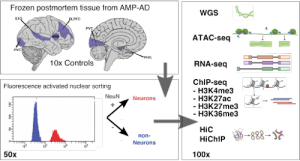
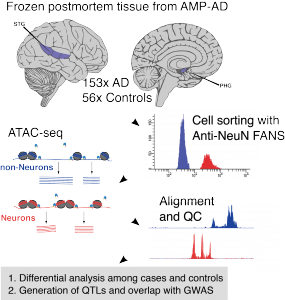
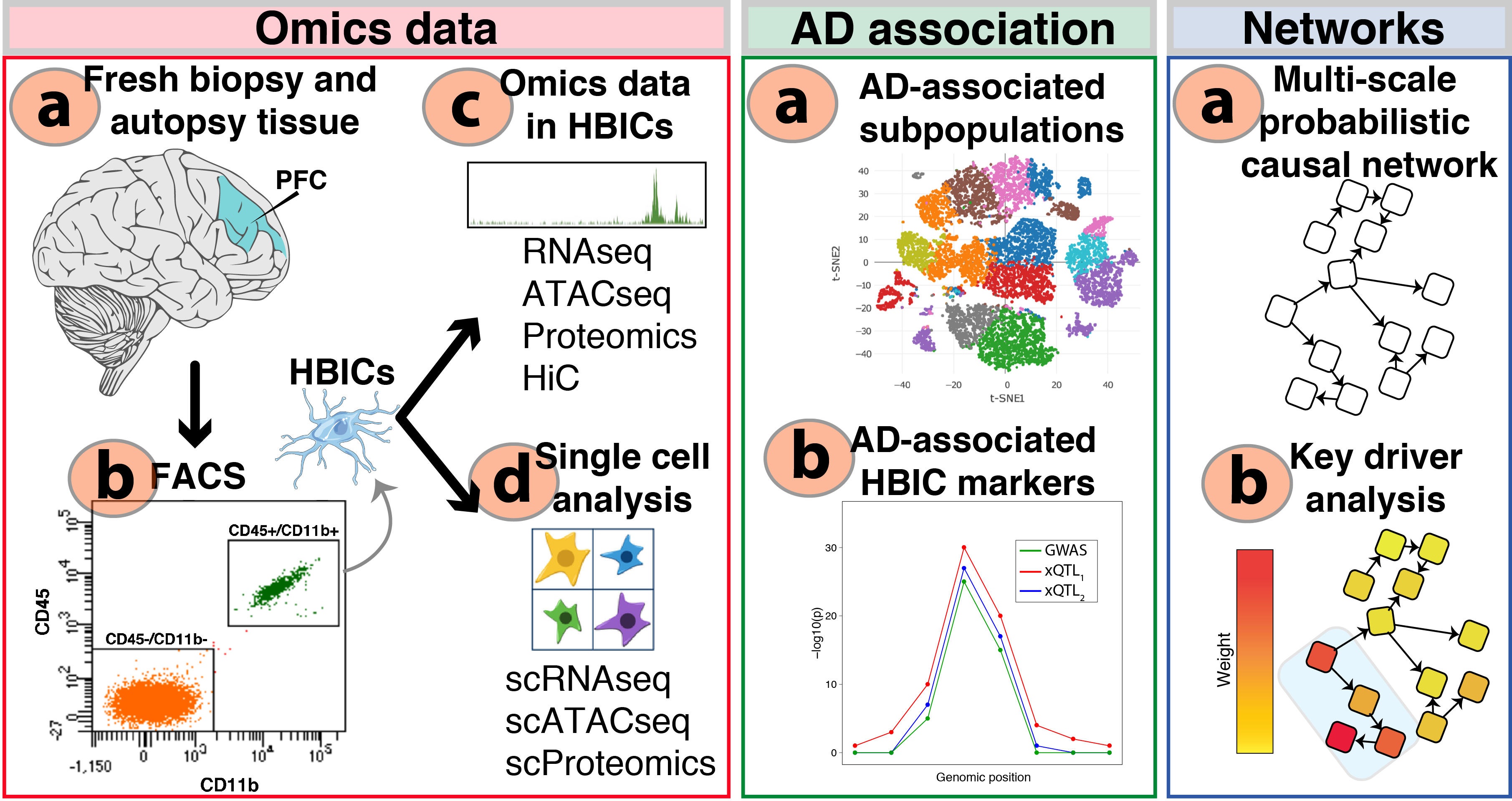
“We generate single cell and cell type-specific multi-scale omics data in the human brain to better understand the genetic architecture and etiopathogenesis of neuropsychiatric diseases.”
Panos Roussos is a Professor of Psychiatry and Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai and Director of the Pamela Sklar Division of Psychiatric Genomics. He is a member of Icahn Institute for Data Science and Genomic Technology and Friedman Brain Institute. He is also a VA/MIRECC Research Physician at the James J. Peters VA Medical Center. He received his medical and doctorate degrees from the University of Crete in Greece and he completed his residency in Psychiatry (research track) at Icahn School of Medicine at Mount Sinai followed by a MIRECC research fellowship in schizophrenia. His early research focused on the genetic exploration of intermediate cognitive phenotypes, including the prepulse inhibition of the startle reflex in human subjects and restoration of deficits using a pharmacogenomic approach. During his residency in psychiatry (Physician-Scientist Research Track) at Icahn School of Medicine at Mount Sinai, he worked on human postmortem studies by integrating genomics with gene expression and gene network approaches. His research focuses on the integration of high-dimensional data, such as genomic, epigenomic, and transcriptomic, using advanced biostatistical methods in order to identify some of the mechanisms through which risk genetic variants increase the risk for neuropsychiatric diseases.

Panos Roussos M.D. Ph.D.
Professor at the Department of Genetics and Genomics Sciences and the Department of Psychiatry
Director of the Pamela Sklar Division of Psychiatric Genomics
Icahn Institute for Data Science and Genomic Technology
Friedman Brain Institute
Icahn School of Medicine at Mount Sinai
Projects
Overview
Functional characterization of non-coding variants for Alzheimer’s disease
Multi-scale analysis in schizophrenia and bipolar disorder
Large-scale transcriptome and epigenome association analysis across multiple traits
Supported by Veterans Administration (Merit BX004189).
Precision Medicine refers to the customization of medical treatment to the individual characteristics of each patient. The Million Veteran Program (MVP) provides a unique opportunity to perform large-scale genome-wide association studies (GWAS) across multiple traits and diseases towards the successful application of Precision Medicine. While well powered GWAS have identified multiple risk variants, due to their small effect sizes there has been limited conclusive findings on the genetic factors contributing to complex traits. In addition, the majority of common risk variants are within non-coding regions of the genome and, as such, the functional relevance of most discovered loci remains unclear. Our group and others have shown that a large proportion of phenotypic variability in disease risk can be explained by regulatory variants, i.e. genetic variants that affect epigenetic mechanisms and the expression levels of genes. The study of gene expression and epigenome changes directly in the MVP samples is not feasible as such data are not available.
To overcome these limitations, we propose to apply a machine learning approach that leverages existing molecular data (unrelated to MVP) as a reference panel to directly impute multi-tissue and genome-wide gene expression and epigenome profiles in MVP samples using existing MVP genotypes. As reference panel, we will use large-scale datasets with genotyping and molecular profiling that our group and others have generated, including, but not limited to, the PsychENCODE project, CommonMind Consortium and Accelerating Medicine Partnership for Alzheimer’s Disease. Imputed MVP gene expression and epigenome data provides a powerful cohort to “translate” genetic findings to the dysregulation of specific molecular pathways across multiple traits that will enhance drug discovery.

The 3D genome in transcriptional regulation across the postnatal life span
Understanding the role of human brain immune cells in Alzheimer Disease vulnerability
Multiregional assessment of gene expression and chromatin accessibility in human brain tissue
Awards & Honors
2017-: Member , American College of Neuropsychopharmacology
2016: Presidential Early Career Awards for Scientists and Engineers, White House
2015: Elected to attend and Awarded Funds, Charleston Conference on Alzheimer’s Disease
2013: Best Research Paper, New York State Psychiatric Association Scientific Paper Contest
2013: Travel Award , International Congress on Schizophrenia Research
2013-: Associate Member , American College of Neuropsychopharmacology
2012: Domestic Travel Fellowship Award, Society of Biological Psychiatry
2012: Research Colloquium for Junior Investigators – Travel Award , American Psychiatric Association
2012: Best Research Paper, New York State Psychiatric Association Scientific Paper Contest
2012: Travel Award , American College of Neuropsychopharmacology
2012: Early Academic Career Award, APA/Merck
2011: Best Research Paper, New York State Psychiatric Association Scientific Paper Contest
2010: Outstanding Resident Award, National Institute of Mental Health
2010: Travel Award, International Society of Psychiatric Genetics
Active Funding
2023 – 2026: BD² Breakthrough Discoveries for thriving with Bipolar Disorder
Dissection of Bipolar Disorder pathophysiology through integration of human brain multi-omics
2023 – 2028: R01AG082185, National Institute of Health/Aging
The adaptive-innate immune interactome across multiple tissues in Alzheimer’s disease
2021 – 2026: R01AG050986, National Institute of Health/Aging
Higher Order Chromatin and Genetic Risk for Alzheimer’s Disease
2021 – 2025: R01MH125246, National Institute of Mental Health
Multiethnic genomic, epigenomic and transcriptomic fine-mapping and functional validation analysis of schizophrenia and bipolar disorder risk loci
2023 – 2025: Foundation for the NIH (FNIH)
STARNET – a multi-tissue omics resource obtained from living patients with cardiometabolic diseases
2020 – 2025: R01AG065582, National Institute of Health/Aging
Understanding the protective and neuroinflammatory role of human brain immune cells in Alzheimer Disease
2019 – 2024: NIH/NIMH/Jefferson University (Roussos Subaward PI)
mGluR5 hypoactivity is integral to glutamatergic dysregulation in schizophrenia
2021 – 2024: RF1MH128970, National Institute of Mental Health
A regulome and transcriptome atlas of fetal and adult human neurogenesis
2019 – 2024: R01DA047880, National Institute on Drug Abuse (Roussos MPI)
Transcriptome and Epigenome Mapping in Dopamine Neurons from the Opioid Exposed Human Brain
2019 – 2024: R01AG067025, National Institute of Health/Aging
Understanding the molecular mechanisms that contribute to neuropsychiatric symptoms in Alzheimer Disease
2018 – 2024: U01MH116442, National Institute of Mental Health (Roussos MPI)
The 3D genome in transcriptional regulation across the postnatal life span, with implications for schizophrenia and bipolar disorder
2019 – 2024: U01DA048279, National Institute on Drug Abuse (Roussos MPI)
Functional genomic resource and integrative model of dopaminergic circuitry associated with psychiatric disease
2021 – 2023: U01NS125580, National Institute of Neurological Disorders and Stroke
Single-nucleus transcriptome profiling across multiple brain regions in Parkinson’s Disease
Previous Funding
2018 – 2020: Merit BX004189, Veterans Administration (Roussos PI) Large-scale transcriptome and epigenome association analysis across multiple traits.
2017 – 2020: R01AG057440, National Institute of Aging (Roussos M-PI) Towards a comprehensive signaling pathway map of parahippocampal vulnerability in Alzheimer’s Disease.
2017 – 2019: Boehringer Ingelheim, LTD (Roussos PI) Multiregional assessment of cell type specific epigenome and transcriptional profiling in human brain tissue.
2016 – 2021: R01MH109677, National Institute of Mental Health (Roussos PI) Risk genetic variants and cis regulation of gene expression in Bipolar Disorder.
2016 – 2021: R01MH109897, National Institute of Mental Health (Roussos M-PI) Integrated Multiscale Networks in Schizophrenia.
2016 – 2020: R01MH110921, National Institute of Mental Health (Roussos PI) Molecular Profiling of Schizophrenia.
2016 – 2021: Presidential Early Career Awards for Scientists and Engineers (PECASE), White House (Roussos PI)
2015 – 2020: R01AG050986,National Institute of Aging (Roussos PI) Higher Order Chromatin and Genetic Risk for Alzheimer’s Disease.
2015 – 2018: R01MH106056,National Institute of Mental Health (Roussos M-PI) Higher Order Chromatin and Genetic Risk for Schizophrenia.
2013 – 2022: Merit BX002395,Veterans Administration (Roussos PI) Dissecting cis regulation of gene expression in schizophrenia.
2016 – 2017: Research Grant, Charleston Conference on Alzheimer’s Disease
2015 – 2017: NIRG-340998, Alzheimer’s Association
2013 – 2015: Young Investigator Award, National Alliance for Research on Schizophrenia and Depression (NARSAD)
2013 – 2014: Research Grant, American Psychiatric Association (APA)
Contact Us
E-mail
Dr. Panos Roussos
Phone
Dr. Panos Roussos: (212) 824-8982
Dr. John Fullard: (212) 824-9125
Address
Roussos Lab
Icahn School of Medicine at Mount Sinai
1470 Madison Ave
The Leon and Norma Hess Center for Science and Medicine
Floor 9, Room 302, Box 1639
New York, NY 10029
Links
- Center for Disease Neurogenomics
- Icahn School of Medicine at Mount Sinai
- Department of Psychiatry
- Department of Genetics and Genomic Sciences – Icahn Institute for Genomics and Multiscale Biology
- Friedman Brain Institute
- PsychENCODE
- CommonMind Consortium
- Accelerating Medicine Partnership for Alzheimer’s Disease
Lab Location
1470 Madison Avenue
Hess Center 9-302 (Lab), 9-107 (Office)
New York, NY 10029
Mail Address:
1470 Madison Avenue
Box 1639
New York, NY 10029