Cancers arise in many organs and presumably from a particular cell of origin. In our laboratory, we are interested in cell-type identity, or cellular lineage, of the cancer cells.
During the tissue development and regeneration, secreted factors in micro-environment from neighboring cells instruct certain cell types to propagate and populate the tissue by providing a defined set of activation signals. This must be tightly controlled by a contextual match between an activation donor signal and a recipient cell with a proper identity. Upon resulting cell division, cells may be spatially removed from the signal and/or instructed to change its identity, or differentiate, to avoid uncontrolled cell growth. Genetic alterations on signaling molecules can provide continuous activation of a signaling cascade. If the cell that acquired such mutation happens to have a matched identity that can respond to the signal and retain that identity, now with the spatial requirement is lost by autonomous activation, the cells may become tolerated to a continuous cell growth signal. This may partially explain why particular genetic alterations are only observed in restricted tissue types, and why tumors from different tissue types carrying the same genetic mutation respond differently to targeted therapies.
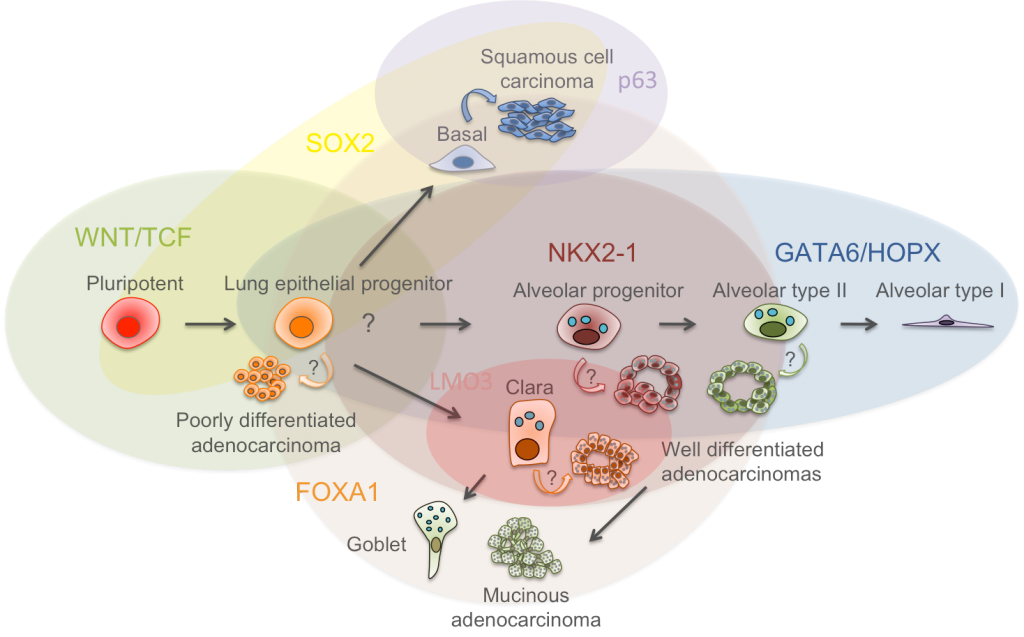
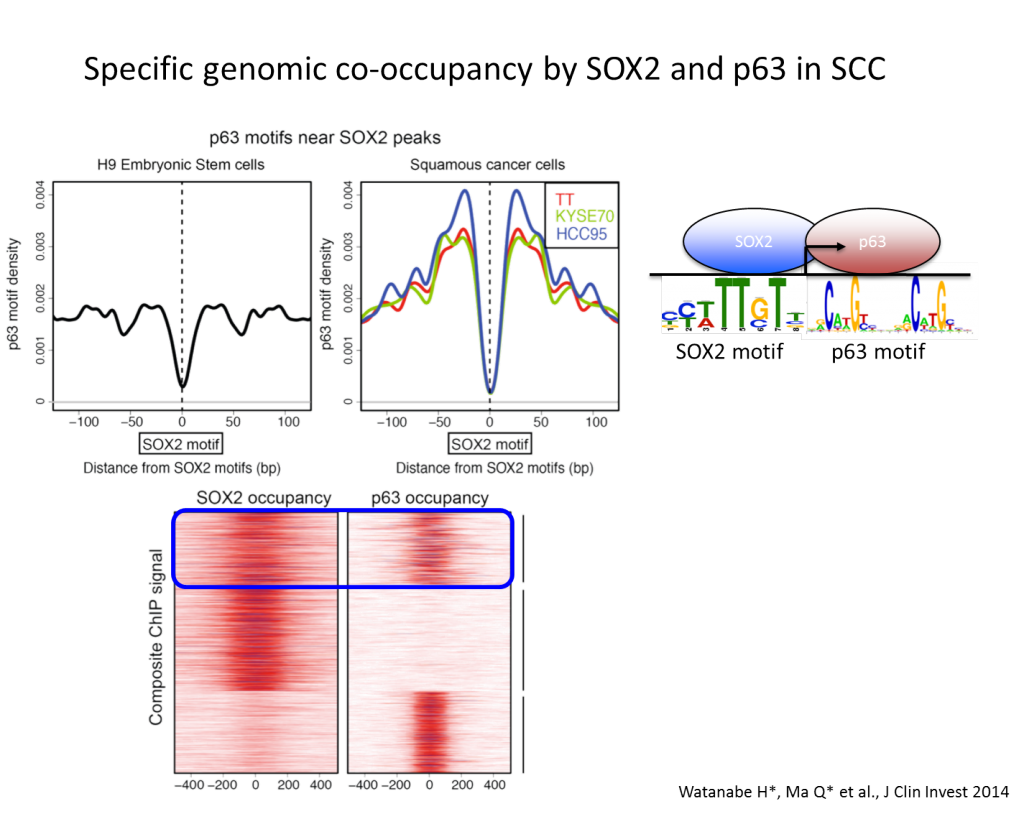
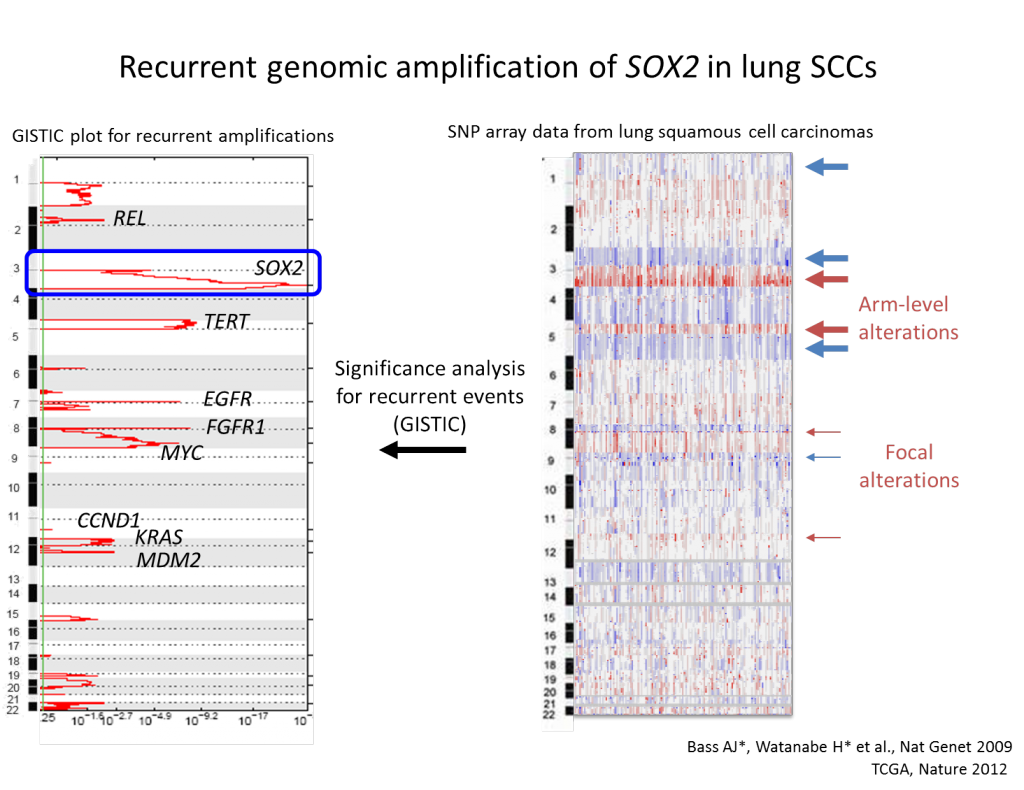
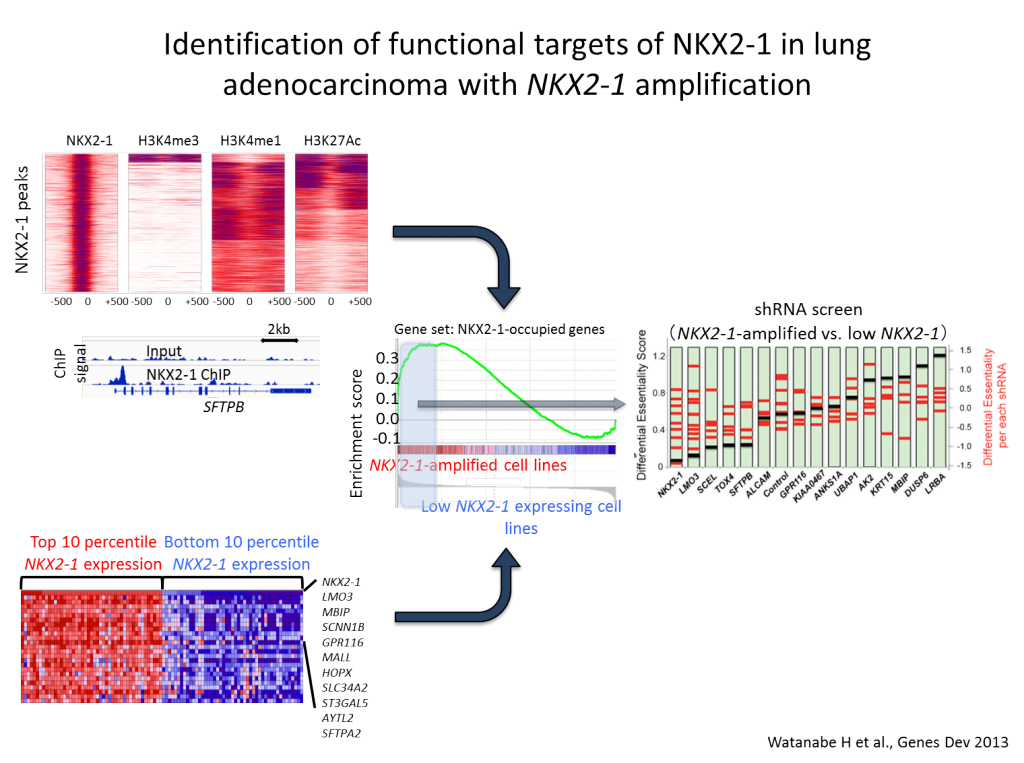
In a regenerative state of the lung, secretory cells have been shown to de-differentiate into stable basal stem cells in response to airway injury, suggesting that even lineage-committed cell types have much higher plasticity than previously appreciated (Tata PR et al, Nature, 2013). Dr. Watanabe proposed a model where lung cancers, in the course of carcinogenesis, lose its cellular plasticity or become fixated to mimic a certain differentiation state that can respond to specific environmental queues (Watanabe, Meyerson, Cancer Cell, 2013). The hypothesis is that a combination of aberrant fixation of lineage such as NKX2-1 amplification and sustained signaling activation such as activating EGFR mutation, if contextually matched, may lead to uncontrolled growth of cells, mimicking transit amplifying cell state. Such notion is supported by the results that discrete depletion of master lineage factor, Nkx2-1, in established lung adenocarcinomas in mice that led to trans-differentiation into mucinous adenocarcinomas rendered increased tumor burden where Kras is activated (Snyder, Watanabe et al., Mol Cell, 2013), whereas similar discrete depletion of Nkx2-1 decreased tumor burden in Egfr-driven lung adenocarcinoma model (Maeda et al., J Clin Invest, 2012). The laboratory pursues invention of sensitively detecting early events on transcriptional programming that lead to fixation of cellular lineage recapitulating transit amplifying cells with a hope to modulate differentiation state of each specific subclass of lung cancers. Understanding of potential roles of differentiation programs in solid tumor carcinogenesis will provide foundation for an additional paradigm in cancer diagnostics and therapeutics.
Our laboratory’s goals is to utilize epigenomics and state-of-the-art genomic technologies to characterize various cellular lineages in normal lung as well as in lung cancer subtypes and identify and elucidate the mechanism of the lineage programs that are aberrantly fixated in lung cancer. We explore the hypothesis that different subsets of lung cancers mimic and depend upon certain differentiation programs, and rely on existence of enhancer clusters for transcription factors driving such programs, and those tumors represent a heterogeneous population of distinct lineages, influenced by genomic contexts and/or microenvironment, as well as influencing the effects of genomic contexts.
We seek to understand 1) what cellular identity the tumors cells may arise from or acquire during carcinogenesis, 2) how they maintain and may depend upon the identity once established, 3) how they may shift identity upon survival pressure from micro-environmental changes or drug treatment, and 4) how mixed populations within a tumor may interact with each other to maintain the tumor as a community over the time of tumor evolution.