Current Research
The mammalian immune system is composed of complex networks of functionally diverse cell types, signaling pathways, regulatory circuits, and molecular mechanisms. Effective host defense depends on coordinated interactions of these numerous components. Recent technological advances have enabled the study of these complex molecular and cellular systems at unprecedented throughput and resolution.
Our laboratory is focused on developing and applying high throughput sequencing and related high dimensional techniques to gain new understandings of immune function in viral infection. Many current projects utilize custom implementations of high throughput single cell RNA-Seq methodologies.
Elucidating antiviral gene networks and effector functions with advanced molecular technologies
As primary innate defenses to viral infection, type I and type III interferons (IFN) establish an antiviral state in both infected and uninfected “bystander” cells through the induction of interferon stimulated genes (ISGs). How the coordinated activities of these hundreds of ISGs confer protection against diverse viruses, and the regulatory circuits that modulate their expression remain poorly understood. Combining molecular biology, physiologically relevant cell culture systems, microfluidics and bioinformatics, our laboratory develops and applies novel methodologies to uncover how single ISGs and functional modules of multiple ISGs restrict viral infection. For example, we have engineered an inducible CRISPR-activation system specialized for the high throughput assessment of ISG functions, which we have used to identify restriction factors for SARS-CoV-2. We are further expanding this and related CRISPR strategies to understand ISG functions in different infection systems and cellular processes. Additional active projects include defining the human airway epithelium responses to IFN and respiratory viruses at single cell resolution, and exploring the different roles of IFN signaling in flavivirus pathogenesis.
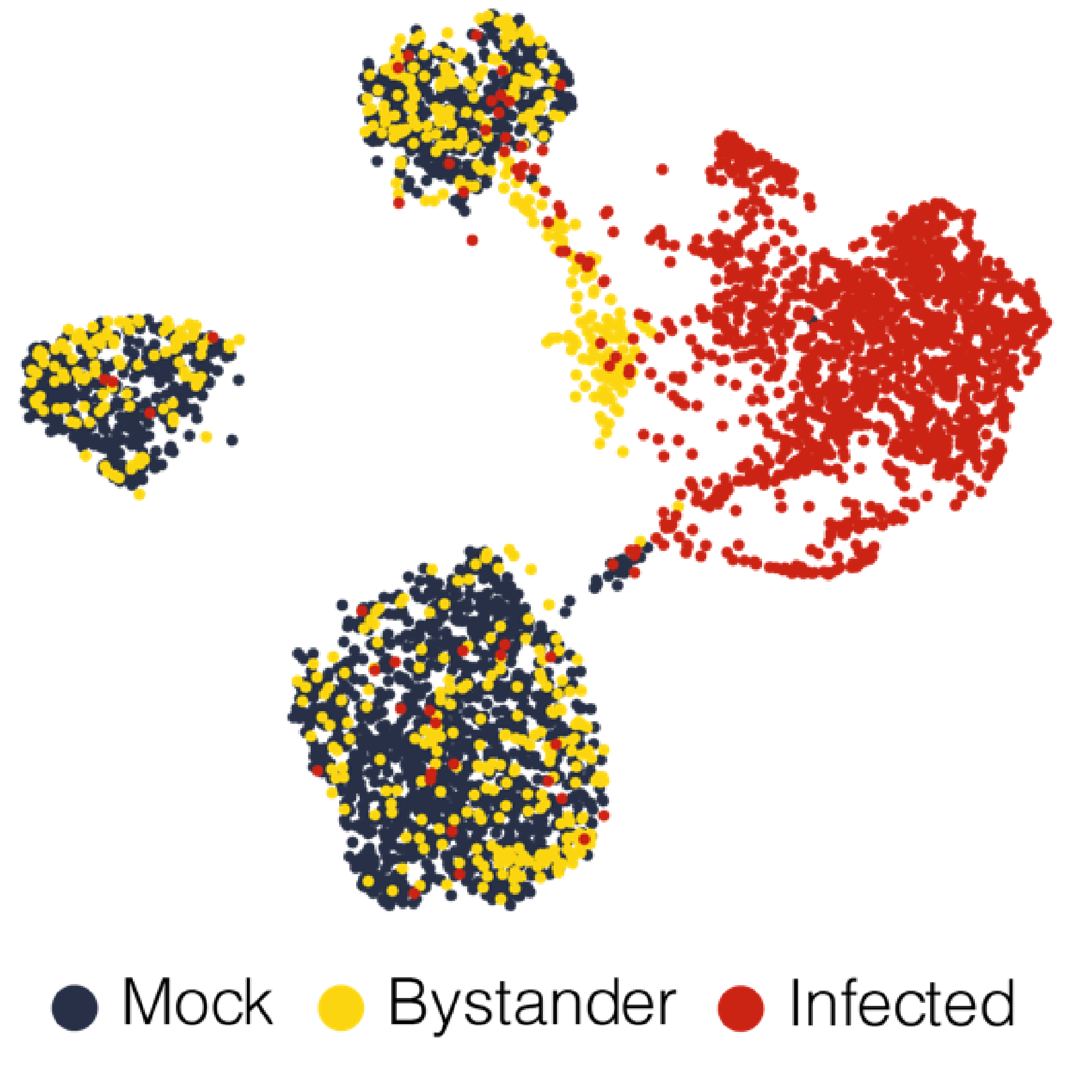
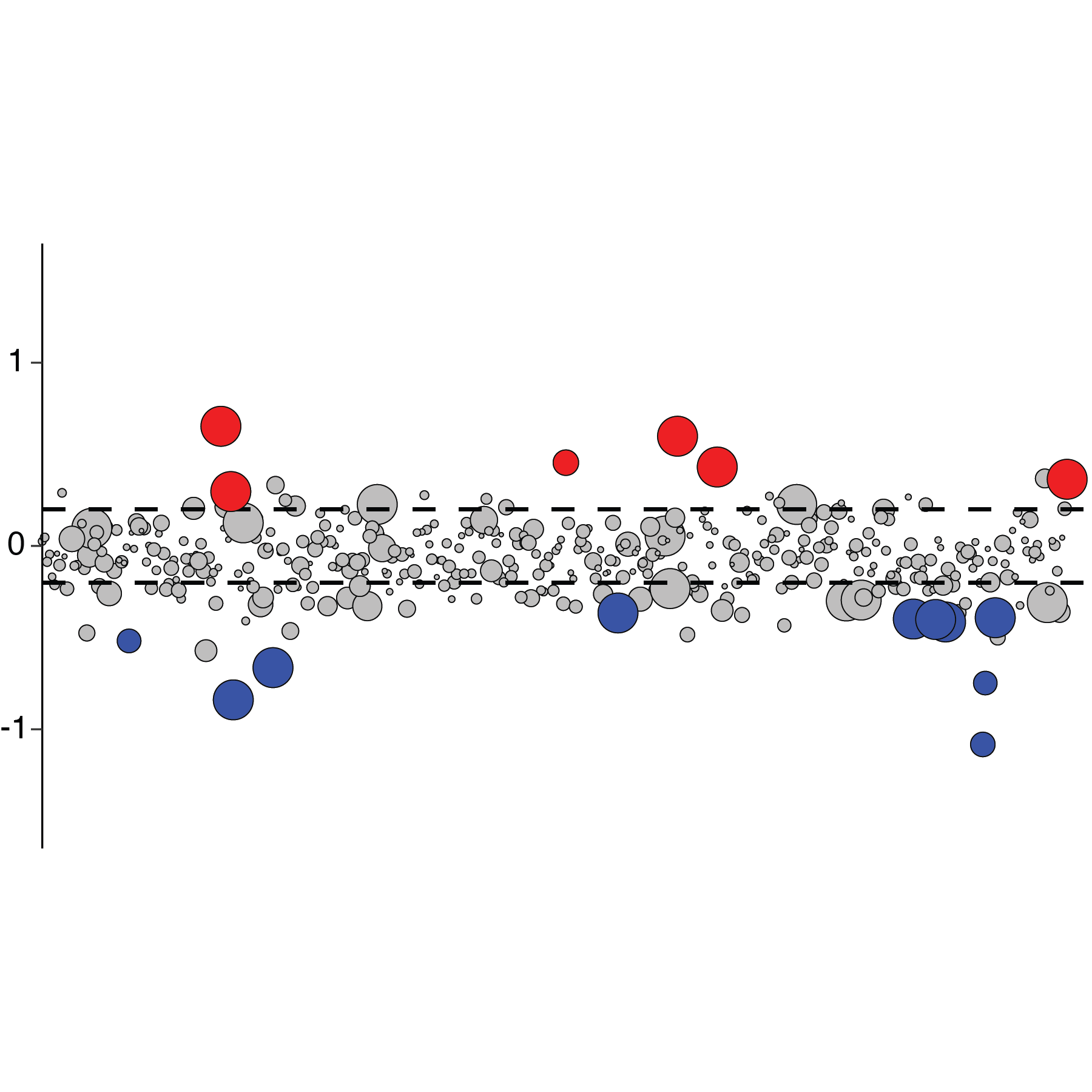
Transcriptomics analysis strategies for diverse aspects of host-virus interactions
Viral infections and corresponding host responses can result in profound transcriptional changes. These may include not only differences in gene expression programs, but also RNA nucleotide modifications and/or alterations in splicing patterns. Moreover, virus-elicited transcriptional changes can vary by cell type, infection status (e.g., directly infected cells versus uninfected “bystander” cells), and genetic composition. RNA-Seq (both “bulk” and single cell) and related methodologies offer powerful tools to investigate these changes and their functional impact at a transcriptome-wide scale. However, studying these molecular processes often requires non-standard molecular biology and analysis strategies. Our laboratory develops and implements tailored transcriptomics workflows to diverse projects exploring the interaction of host cells and viral pathogens. Recent examples include establishing a method for the unambiguous detection of SARS-CoV-2 mRNAs by single cell RNA sequencing, modifying a droplet microfluidics single cell RNA-Seq platform to resolve cell-intrinsic IFN signaling phenotypes in genetic mosaicism, defining the “editome” of the ADAR1 RNA editing enzyme and its impact on the innate antiviral response, and quantifying transcript splice intermediates in virus-susceptible humans with a genetic deficiency in RNA processing.
Single cell RNA-Seq for immunology and virology in “non-traditional” model organisms
Infectious disease research must often rely on “non-traditional” model organisms to best recapitulate features of human pathology and to understand zoonotic agents. However, such research presents challenges, such as relatively limited research tools (e.g. species-compatible immunoreagents, culture systems), and an incomplete knowledge of potentially species-specific cell types. Our laboratory utilizes single cell RNA-Seq strategies for immunology and virology in species for which traditional experimental tools are limited. Examples include characterizing immune cell diversity and host response to an equine hepacivirus in horses, investigating the response to SARS-CoV-2 in the Syrian golden hamster model, and characterizing the immune response to Influenza A virus in ferrets, a medically important model for Influenza pathogenesis and vaccinology.
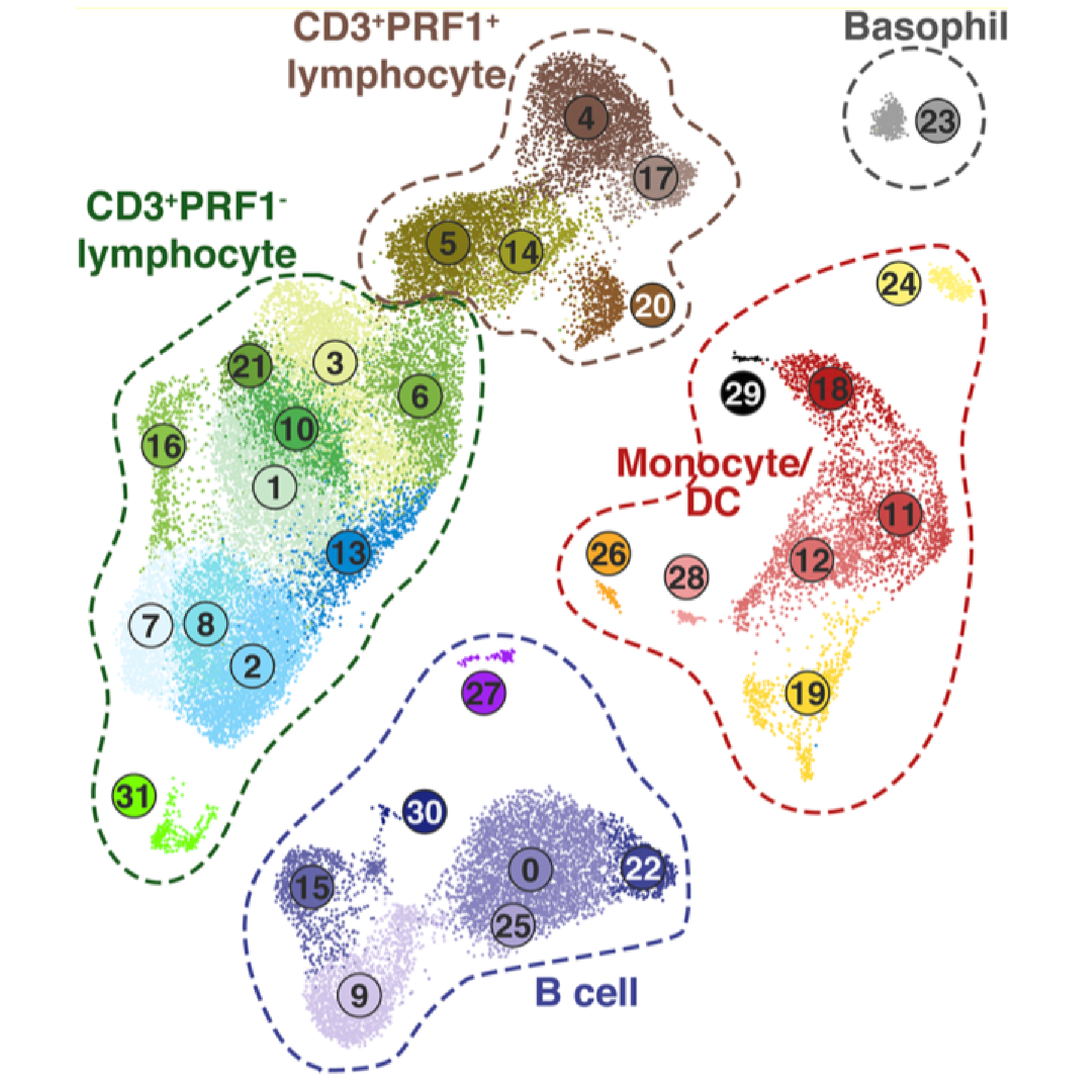
Elucidating antiviral gene networks and effector functions with advanced molecular technologies
As primary innate defenses to viral infection, type I and type III interferons (IFN) establish an antiviral state in both infected and uninfected “bystander” cells through the induction of interferon stimulated genes (ISGs). How the coordinated activities of these hundreds of ISGs confer protection against diverse viruses, and the regulatory circuits that modulate their expression remain poorly understood. Combining molecular biology, physiologically relevant cell culture systems, microfluidics and bioinformatics, our laboratory develops and applies novel methodologies to uncover how single ISGs and functional modules of multiple ISGs restrict viral infection. For example, we have engineered an inducible CRISPR-activation system specialized for the high throughput assessment of ISG functions, which we have used to identify restriction factors for SARS-CoV-2. We are further expanding this and related CRISPR strategies to understand ISG functions in different infection systems and cellular processes. Additional active projects include defining the human airway epithelium responses to IFN and respiratory viruses at single cell resolution, and exploring the different roles of IFN signaling in flavivirus pathogenesis.
Transcriptomics analysis strategies for diverse aspects of host-virus interactions
Viral infections and corresponding host responses can result in profound transcriptional changes. These may include not only differences in gene expression programs, but also RNA nucleotide modifications and/or alterations in splicing patterns. Moreover, virus-elicited transcriptional changes can vary by cell type, infection status (e.g., directly infected cells versus uninfected “bystander” cells), and genetic composition. RNA-Seq (both “bulk” and single cell) and related methodologies offer powerful tools to investigate these changes and their functional impact at a transcriptome-wide scale. However, studying these molecular processes often requires non-standard molecular biology and analysis strategies. Our laboratory develops and implements tailored transcriptomics workflows to diverse projects exploring the interaction of host cells and viral pathogens. Recent examples include establishing a method for the unambiguous detection of SARS-CoV-2 mRNAs by single cell RNA sequencing, modifying a droplet microfluidics single cell RNA-Seq platform to resolve cell-intrinsic IFN signaling phenotypes in genetic mosaicism, defining the “editome” of the ADAR1 RNA editing enzyme and its impact on the innate antiviral response, and quantifying transcript splice intermediates in virus-susceptible humans with a genetic deficiency in RNA processing.
Single cell RNA-Seq for immunology and virology in “non-traditional” model organisms
Infectious disease research must often rely on “non-traditional” model organisms to best recapitulate features of human pathology and to understand zoonotic agents. However, such research presents challenges, such as relatively limited research tools (e.g. species-compatible immunoreagents, culture systems), and an incomplete knowledge of potentially species-specific cell types. Our laboratory utilizes single cell RNA-Seq strategies for immunology and virology in species for which traditional experimental tools are limited. Examples include characterizing immune cell diversity and host response to an equine hepacivirus in horses, investigating the response to SARS-CoV-2 in the Syrian golden hamster model, and characterizing the immune response to Influenza A virus in ferrets, a medically important model for Influenza pathogenesis and vaccinology.
