- What is Craniosynostosis?
- Craniosynostosis Network
- Network Organization
- Network Directory
- News & Resources
- FAQs
- Publications
- Contact us
Craniosynostosis Network
Craniosynostosis Network is an organized and synergistic research program to understand the genetic basis and developmental mechanism of craniosynostosis.
The objective of Craniosynostosis Network is to identify and functionally validate genes and biological pathways contributing to common forms of cNSC and to the related continuum of craniofacial phenotypes.
Aim: To increase our understanding of the genetic basis and developmental dynamics of craniosynostosis phenotypes through a collaborative investigation of cell function in mouse models and in humans with these conditions, and discovery of new genetic variants causative for coronal craniosynostosis. This will be accomplished through collaborative investigation of the aims of three Projects and two Cores:
Project I: From Skull Shape to Cell Activity in Coronal Craniosynostosis
Project II: Genomic Approaches to Coronal Nonsyndromic Craniosynostosis
Project III: Systems Biology of Bone in Coronal Nonsyndromic Craniosynostosis
Core A: Administrative Core
Core B: Molecular/Analytic Core
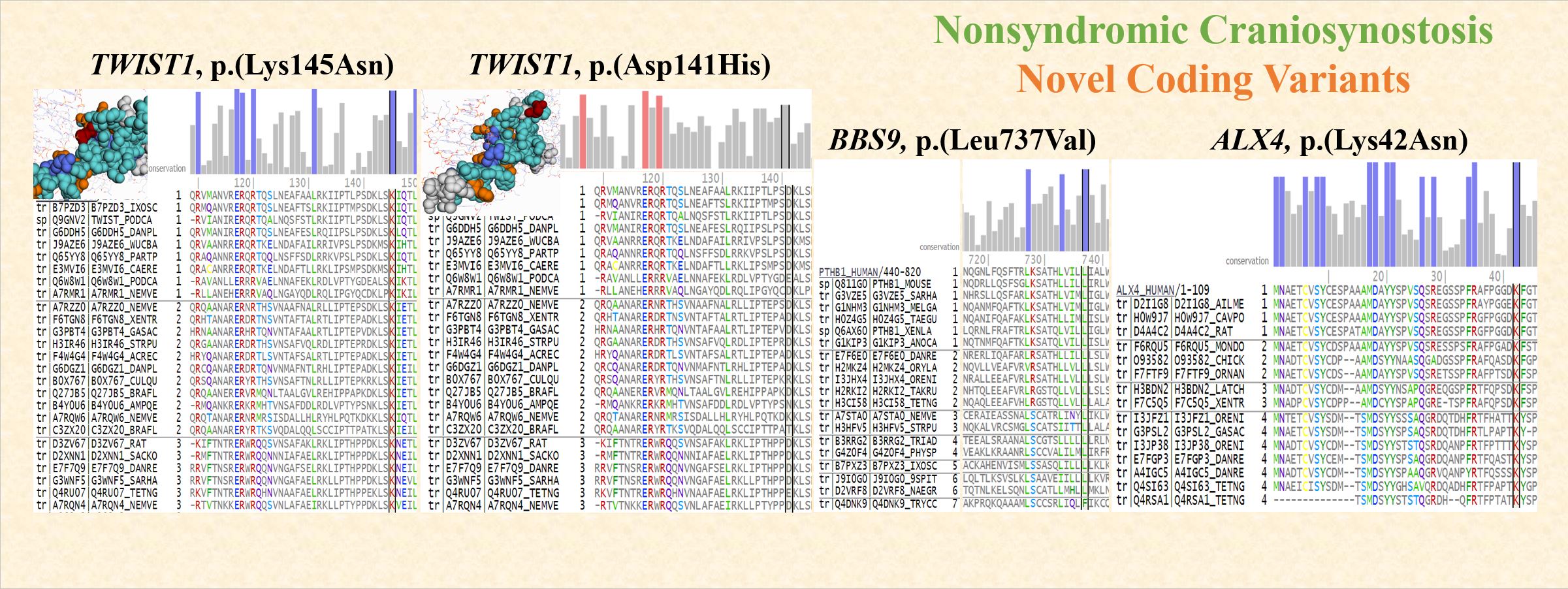 FIGURE: Functional consequences of novel mutations: a TWIST1 p.(Lys145Asn) acts as a transcriptional regulator for cranial suture patterning and fusion. The mutation affects the evolutionarily conserved residue and one of the key binding site residues. Functional impact predicted by Mutation Assessor is “Medium”. b TWIST1 p.(Asp141His) mutation affects one of the high-scoring specificity residues, i.e., residues conserved within protein subfamilies. Functional impact predicted by Mutation Assessor is ”Medium”. c BBS9 p.(Leu737Val) is required for proper BBSome complex assembly which is required for ciliogenesis. The mutation affects the evolutionarily conserved residue. Functional impact predicted by Mutation Assessor is “High”. d ALX4 p.(Lys42Asn) encodes a paired-like homeodomain transcription factor expressed in the mesenchyme of developing bones. The mutation affects the evolutionarily conserved residue. Reference: Sewda et al (2019); https://www.nature.com/articles/s41390-019-0274-2
FIGURE: Functional consequences of novel mutations: a TWIST1 p.(Lys145Asn) acts as a transcriptional regulator for cranial suture patterning and fusion. The mutation affects the evolutionarily conserved residue and one of the key binding site residues. Functional impact predicted by Mutation Assessor is “Medium”. b TWIST1 p.(Asp141His) mutation affects one of the high-scoring specificity residues, i.e., residues conserved within protein subfamilies. Functional impact predicted by Mutation Assessor is ”Medium”. c BBS9 p.(Leu737Val) is required for proper BBSome complex assembly which is required for ciliogenesis. The mutation affects the evolutionarily conserved residue. Functional impact predicted by Mutation Assessor is “High”. d ALX4 p.(Lys42Asn) encodes a paired-like homeodomain transcription factor expressed in the mesenchyme of developing bones. The mutation affects the evolutionarily conserved residue. Reference: Sewda et al (2019); https://www.nature.com/articles/s41390-019-0274-2
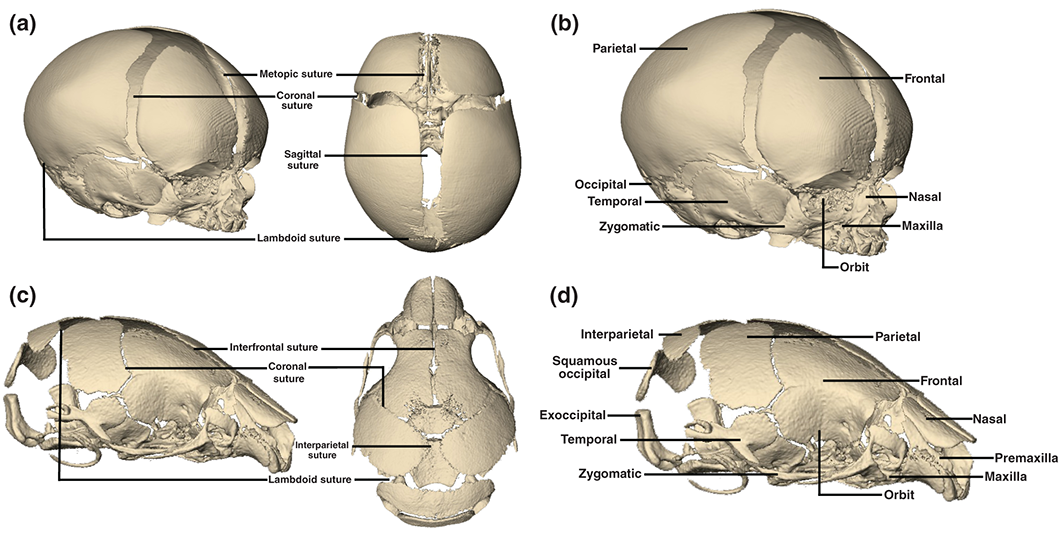 Figure: 3D reconstructions of CT images of a neonatal human cranium (a and b) and newborn mouse cranium (c and d) illustrating corresponding cranial bones and sutures of the two species. Reference: Flaherty et al (2016); https://onlinelibrary.wiley.com/doi/full/10.1002/wdev.227
Figure: 3D reconstructions of CT images of a neonatal human cranium (a and b) and newborn mouse cranium (c and d) illustrating corresponding cranial bones and sutures of the two species. Reference: Flaherty et al (2016); https://onlinelibrary.wiley.com/doi/full/10.1002/wdev.227
