Research Summary
The Mullins Lab is part of the Department of Psychiatry, Department of Genetics and Genomic Sciences, and the Charles Bronfman Institute for Personalized Medicine. Our research involves large-scale genomic studies of psychiatric disorders. We use statistical and bioinformatics approaches to identify risk loci, determine relevant tissues, cell-types and pathways, prioritize causal genes and potential therapeutic targets, and develop genetic risk predictors. We work primarily on bipolar disorder and suicidality, two heritable psychiatric illnesses for which improved treatments, preventions and diagnostics are urgently needed.
Current Projects
Genomics of bipolar disorder in the Psychiatric Genomics Consortium
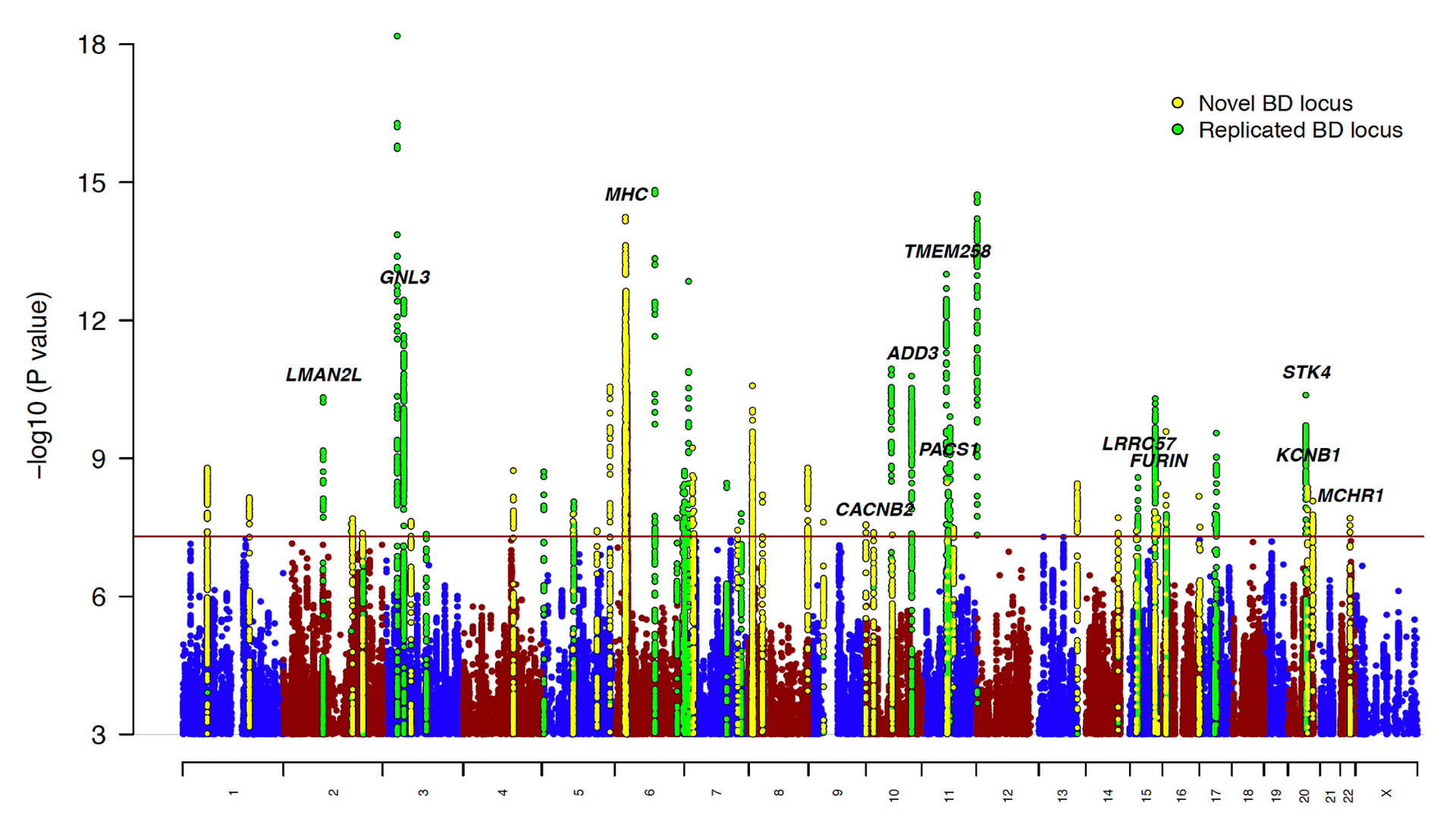
We are key contributors to the Bipolar Disorder Working Group of the Psychiatric Genomics Consortium. Our latest genome-wide association study (GWAS) of bipolar disorder (BD) included ~42,000 BD cases of European descent, and implicated 64 genomic risk loci (Mullins et al., Nat Genet, 2021). The study found enrichment of BD risk alleles in genes expressed in neurons and in genes encoding targets of antipsychotics, calcium channel blockers and antiepileptics. Integrating eQTL data prioritized genes linked to BD via gene expression, including druggable targets. Causal inference analysis highlighted relationships between BD, smoking, alcohol consumption and sleep disturbances. In the PGC, we are now working to increase ancestral diversity in BD GWAS via global outreach, study the spectrum of BD in the population in collaboration with 23andMe, and dissect the genetic and clinical heterogeneity of BD.
Statistical and functional fine-mapping of genetic risk loci for psychiatric disorders
While hundreds of genetic risk loci for psychiatric disorders have been identified through GWAS, the causal variants and genes within most of these loci remain unknown. Statistical and functional fine-mapping is a rapid, scalable and cost-effective way to prioritize likely causal genes and variants that are promising candidates for experimental validation. In this project, we are developing a comprehensive statistical and functional fine-mapping pipeline optimized for psychiatric disorders, which leverages a suite of statistical and functional fine-mapping methods, and ‘omics data from biologically relevant tissues, cell-types, and developmental timepoints. This pipeline is currently being applied to fine-map GWAS risk loci for bipolar disorder.
Genomics of suicidality in the Psychiatric Genomics Consortium
Dr. Mullins is Co-chair of the Suicide Working Group of the Psychiatric Genomics Consortium. As a Postdoctoral Fellow, she co-founded the International Suicide Genetics Consortium (ISGC) to conduct powerful GWAS of suicide outcomes, by combining data worldwide. The ISGC published a GWAS of ~30,000 individuals with a history of suicide attempt, versus >500,000 controls. This study identified the first replicable genome-wide significant locus for suicide attempt, and demonstrated the common variant genetic etiology of suicide attempt after conditioning on related psychiatric disorders (Mullins et al., Biol Psychiatry, 2022). Now, as the Suicide Working Group of the PGC, we are conducting a comprehensive interrogation of the genetic etiologies of suicide attempt, suicide, and suicidal ideation, to identify their underlying biology, modifiable risk factors, and potential therapeutic targets.
Risk prediction of suicide outcomes in hospital populations
We are developing clinical and genetic risk predictors for suicide outcomes in hospital electronic health record (EHR)-linked biobanks. In collaboration with the Coon Lab at the University of Utah, and colleagues from the New York City Clinical Data Research Network, we are applying machine learning algorithms identify individuals at risk of suicide or suicide attempt in EHRs, including the Mount Sinai BioMe biobank. Information from physician notes and other clinical data will be combined with polygenic risk scores trained based on large-scale GWAS, to develop risk prediction models for suicide attempt or suicide, with the goal of stratifying high-risk groups of patients for intervention, treatment and prevention.
Delineating the network effects of mental disorder-association variants using convex optimization methods
This new multidisciplinary project is a collaboration with Dr. David Knowles’s Lab at New York Genome Center and Columbia University. Psychiatric disorders are highly comorbid, have substantial diagnostic overlap, and share clinical features and genetic etiology. In this project, new computational methods being developed by the Knowles Lab will be applied to: 1) determine the shared and distinct genetic components across neuropsychiatric disorders and their subphenotypes, via latent factor analyses applied to genome-wide association study (GWAS) results, 2) infer the gene regulatory and causal networks underlying these components by integrating ‘omics data from brain tissues and cell-types via network causal inference analysis, and 3) uncover how common and rare genetic variants disrupt these networks using a variety of network-based approaches for modeling mutational data.
Team

Niamh Mullins, PhD
Associate Professor
Principal Investigator
niamh.mullins@mssm.edu

Maria Koromina, PhD
Instructor

Sarah Colbert, BA
PhD Student

Brian Fennessy, MSc
Bioinformatician II

Shane O’Connell, PhD
Postdoctoral Fellow

Lucas Toshio Ito, BSc
Visiting PhD Student
Past Members
Shane Crinion, Visiting PhD Student, University of Galway
Renata Gonzalez Chong, Clinical Research Coordinator II
Collaborators
Alexander Charney, MD, PhD (Icahn School of Medicine at Mount Sinai)
Hilary Coon, PhD (University of Utah)
Anna Docherty, LP, PhD (University of Utah)
David Knowles, PhD (New York Genome Center, Columbia University)
Towfique Raj, PhD (Icahn School of Medicine at Mount Sinai)
Douglas Ruderfer, PhD (Vanderbilt University)
Bipolar Disorder Working Group of the Psychiatric Genomics Consortium
Suicide Working Group of the Psychiatric Genomics Consortium
Publications
Complete List of Published Work in Google Scholar and MyNCBI Bibliography.
Funding
Active
NIMH R01MH132733, “Establishing the Suicide Working Group of the Psychiatric Genomics Consortium to elucidate the genetics and biology of suicide outcomes”, PI Mullins
NIMH R01MH124839, “3/7 Psychiatric Genomics Consortium: Advancing Discovery and Impact”, MPIs Mullins, Huckins, Andreassen and Iyegbe
NIMH R01MH130879, “Delineating the network effects of mental disorder-associated variants using convex optimization methods”, PI Knowles (New York Genome Center), Co-I / MSSM PI Mullins
NIMH R01MH123451, “Latino Ancestry Genomic Psychiatry Cohort”, MPI: Pato (Rutgers), Bigdeli, Fanous, and Pato, Co-I / MSSM PI Mullins
NIMH R01MH123489, “Predicting suicide death using EHR and polygenic risk scores”, PI Coon (University of Utah), Co-I / MSSM PI Mullins
International OCD Foundation, “Dissecting the temporal and causal relationships between OCD and bipolar disorder”, PI Grice, Co-I Mullins
Recently Completed
Baszucki Brain Research Fund and Milken Institute Center for Strategic Philanthropy, “Statistical and functional fine-mapping of bipolar disorder genetic risk loci”, PI Mullins
NARSAD Young Investigator Award, Brain & Behavior Research Foundation, “Identifying genetic associations with suicide attempt and characterizing the genetic profiles of high-risk patients in hospital populations”, PI Mullins
Awards
2023, Early Career Investigator Oral Presentation Award, International Society of Psychiatric Genetics, “Novel insights into the genetic etiology of bipolar disorder from a multi-ancestry genome-wide association study by the Psychiatric Genomics Consortium”, awarded to Maria Koromina
2023, Early Career Investigator Oral Presentation Award, International Society of Psychiatric Genetics, “Genome-wide association studies of suicidal thoughts and behaviors: An update from the PGC Suicide Working Group”, awarded to Sarah Colbert
2023, Somerfeld-Ziskind Research Award, Society of Biological Psychiatry, “Dissecting the Shared Genetic Architecture of Suicide Attempt, Psychiatric Disorders, and Known Risk Factors. Biol Psychiatry, 2022”, awarded to Niamh Mullins and Jay Kang
2020, Gershon Paper of the Year Award, International Society of Psychiatric Genetics, “GWAS of Suicide Attempt in Psychiatric Disorders and Association With Major Depression Polygenic Risk Scores. Am J Psychiatry, 2019”, awarded to Niamh Mullins
2019, Early Career Investigator Oral Presentation Award, International Society of Psychiatric Genetics, “Insights into the genetics of suicide attempt from the International Suicide Genetics Consortium”, awarded to Niamh Mullins
Positions
Dr. Mullins is a Graduate Research Training Faculty Member of the Graduate School of Biomedical Sciences, and the Mullins Lab accepts graduate students.
We also have open positions for Postdoctoral Fellows or staff scientists to work on ongoing projects. Feel free to get in touch to discuss potential opportunities!
We provide a vibrant work environment, with access to large-scale genomic datasets, high performance scientific computing facilities, seminar series, training workshops, local, national, and international conferences, funding opportunities, and mentorship resources. The Mullins Lab is committed to fostering a culture that is welcoming and inclusive to diverse groups of individuals, and is passionate about supporting the development of team members scientifically and professionally. We offer competitive salaries and relocation assistance. The Icahn School of Medicine at Mount Sinai offers visa sponsorship, medical benefits, subsidized child care resources, and subsidized apartment rentals in Manhattan to Postdoctoral Fellows and their families.
