Research
The main focus of our lab is the study of Wnt/Frizzled-planar cell polarity (PCP) signaling and associated regulatory and signaling specificity mechanisms. Almost all cells display a polarity and epithelial cells are polarized in two axes, apical-basolateral polarity enables tissues to perform vectorial functions, like directed secretion of specialized components, and planar polarity (PCP in the plane of an epithelium) is often associated with general tissue and organ patterning. Most cells are polarized via PCP with respect to the body axes. This is not only very evident in Drosophila but is a widespread feature of epithelia in all vertebrates including humans. For example, tissues presenting obvious PCP patterning features include skin development, inner ear sensory cells, and kidney development, or also directed movement and intercalation of mesenchymal cell populations during gastrulation and neurulation. To study the mechanisms of Wnt/Frizzled PCP patterning we are using a combination of Drosophila in vivo studies, cell culture experiments, and biochemistry and through collaborations we confirm our results in vertebrate models like zebrafish or mouse.
Featured Projects

Marek Mlodzik, PhD
marek.mlodzik@mssm.edu
Professor & Chair | Cell, Developmental & Regenerative Biology
Professor | Ophthalmology
Professor | Oncological Sciences
Wnt/Frizzled PCP signaling
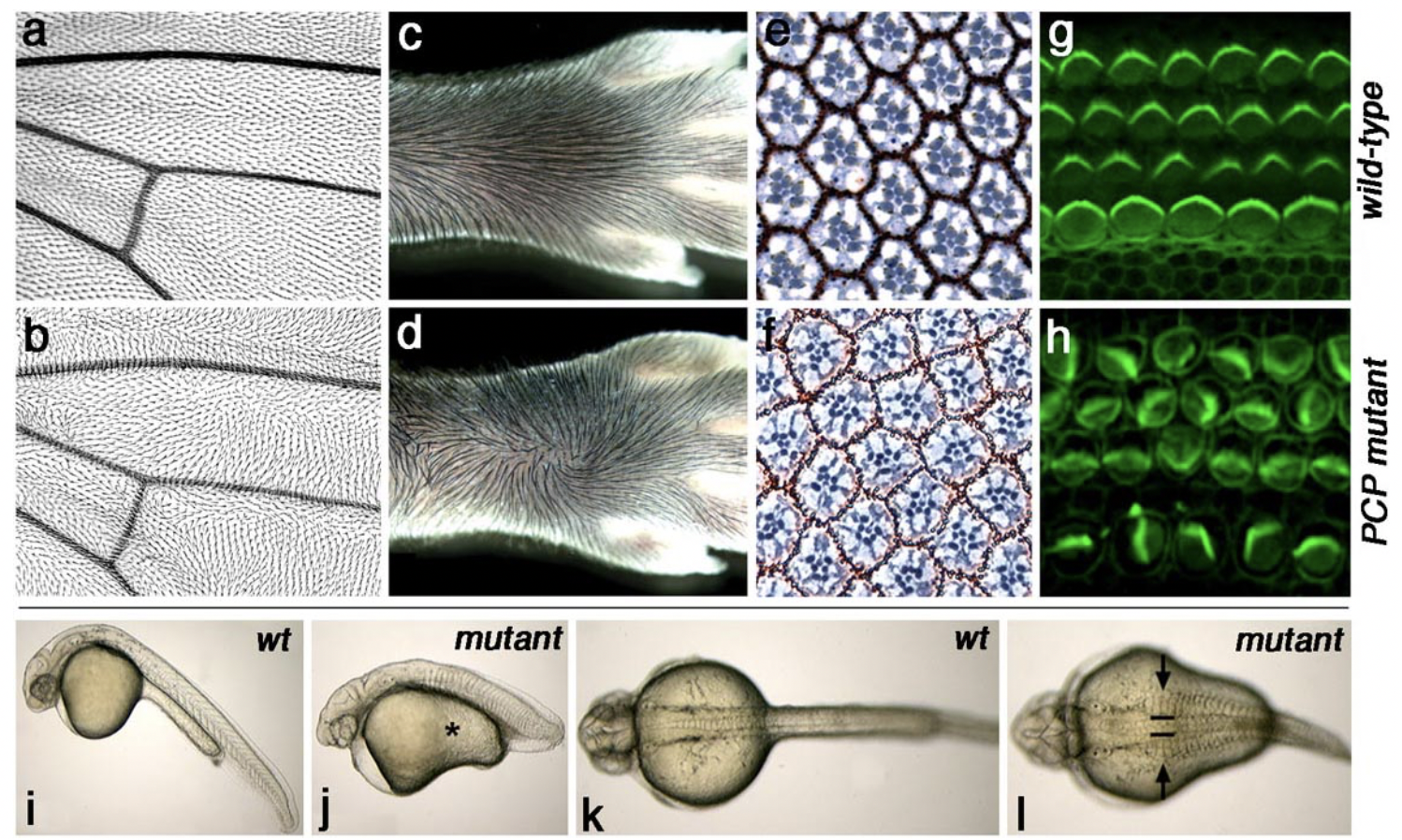
The PCP core genes are required for correct establishment of tissue polarity in all tissues analyzed (including neuronal cells). See examples of PCP defects in Drosophila wings and eyes, mammalian skin and inner ear (mouse), and vertebrate body patterning (zebrafish) in figure above.

A key feature of PCP signaling is the resulting asymmetric PCP protein localization (as schematized above). For example in the wing in the proximo-distal axis and in the eye in the dorso-ventral axis.
The lab focuses in particular on the molecular understanding of the regulation of Wnt/Fz signaling pathway(s) that are key regulators of the process and its interactions with other signaling pathways involved, for example Notch signaling in the fly eye or Fat/Dachsous signaling in general PCP establishment.
Our efforts to understand and molecularly dissect the mechanisms of PCP formation are focusing on the eye and wing in Drosophila as these tissues provide unique models to study specific aspects of PCP establishment. In the wing, PCP is reflected in the choice of the site at which an actin-based out-growth (“hair ”) initiates in each cell and the direction the hair points, with PCP controlling cytoskeletal organization. In the eye, PCP is reflected in the mirror-symmetric arrangement of individual facets or ommatidia relative to the dorso-ventral midline and associated distinct cell fate decisions and signaling, and hence PCP is controlling here a nuclear signaling pathway. Mutations in PCP genes result in the loss of wing hair orientation and ommatidial arrangements, respectively. A key feature of PCP signaling is the resulting asymmetric localization of the core PCP proteins themselves. For example in the wing in the proximo-distal axis and in the eye in the dorso-ventral axis. The underlying signaling pathway(s) and molecular features are conserved throughout evolution and regulate related developmental aspects of coordinated cellular polarization in humans during development and disease.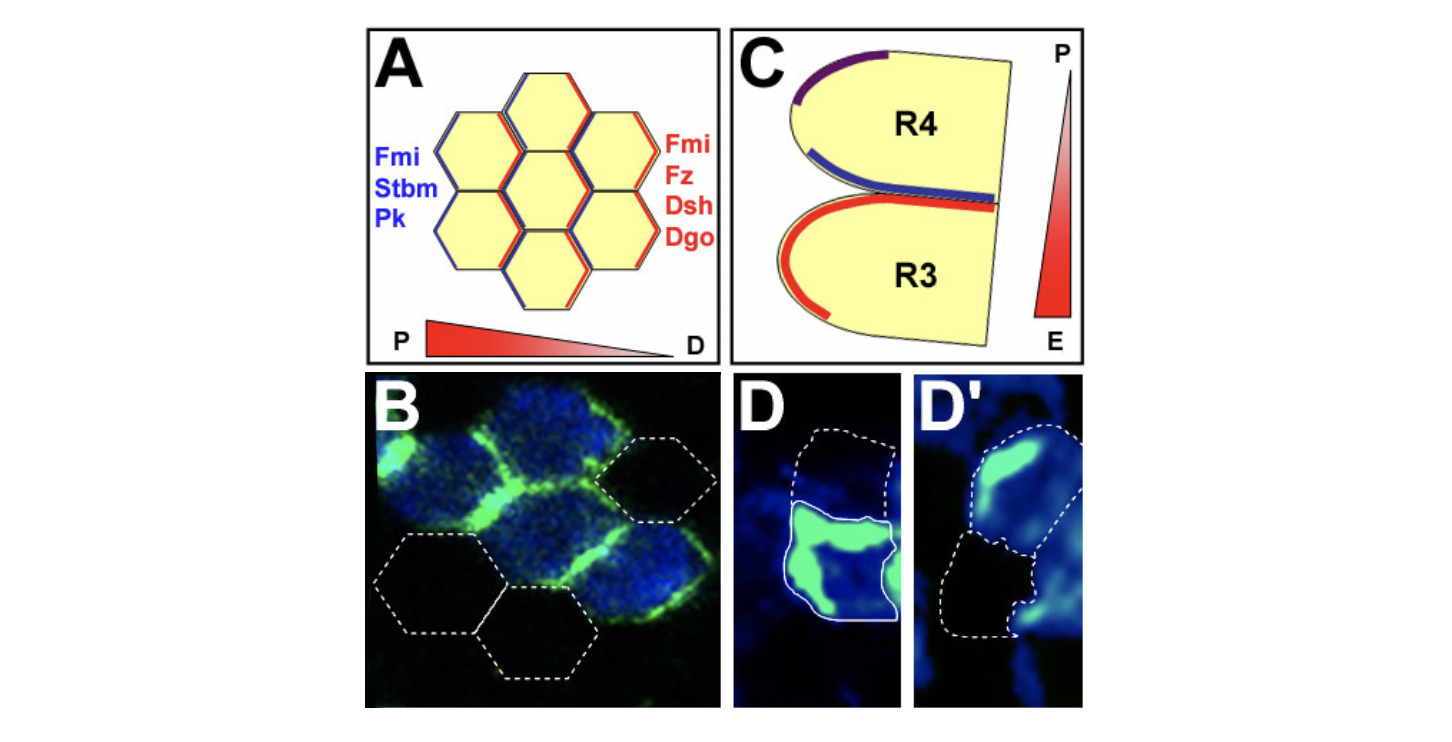
Core PCP protein localization in Drosophila wing and eye cells. (A,B) PCP proteins are asymmetrically localized to proximal and distal cell membranes in pupal wing. Orientation of the gradient indicates the presumed Fz activity. (A) Schematic of localization of PCP proteins; hexagons represent single cells. (B) Confocal image with distal localization of Diego (green), as an example. White outlines: cells not expressing GFP-Diego. (C,D) Eye disc: core PCP proteins are asymmetrically localized. (C) Schematic of R3/R4 cells (colors as in A; purple: colocalization of all factors).
Wnt/Frizzled β-catenin signaling and Wnt-signaling specificity
A second focus of the lab is to understand how signaling specificity is achieved between the canonical Wnt/b-catenin pathway and Wnt/Fz-PCP signaling. The two pathways bifurcate at the level of Dishevelled (Dsh) and our efforts are currently concentrated on dissecting the pathway specific modifications of Dsh and the interaction(s) of Dsh with the Fz receptor complexes. Pathway specific Dsh membrane recruitment and Dsh phosphorylation are being studied in through functional genomics in cell culture assays, biochemical dissection, and in vivo studies. We have identified several genes that help in switching from on pathway to the other and that affect Dsh phosphorylation in distinct ways.
In parallel, we have identified a novel role for IFT-A ciliary proteins in the Wnt/b-catenin pathway acting downstream of b-catenin stabilization and upstream of transcriptional events. This IFT-A function is independent of the ciliary role of IFTs and is cytoplasmic and conserved in mammals. We have demonstrated that IFT-A and Kinesin 2 act together as a complex that binds β-catenin and microtubules and is required for nuclear translocation of b-catenin. Activation of Wnt-signaling in IFT-A or Kinesin 2 mutants fails to activate Wg/Wnt targets, due to markedly reduced nuclear β-catenin localization, although high levels of cytoplasmic β-catenin are detected. Many questions remain however: for example (i) How is the Kinesin-2/IFT-A complex promoting nuclear localization? Preliminary data suggest it is by movement along microtubules, but this needs to be confirmed and the associated exciting cell biology dissected; and (ii) our preliminary data further suggest that there is an antagonistic factor that competes with IFT140 binding to β-catenin to ‘inhibit’ its nuclear translocation. We are addressing these exciting questions.
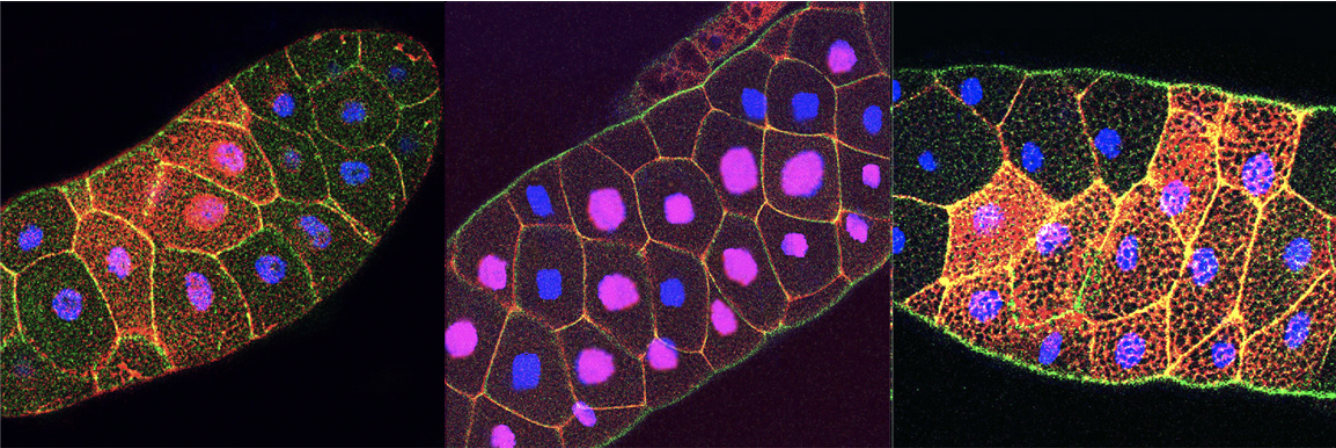
Figure: Nuclear localization of β-catenin (red) after Wnt-induction (left panel) or in axin-/- mutants (middle panel) is blocked in IFT140 mutants (right panel). E-cadherin is in green and DAPI in blue
Cell motility as regulated by Wnt-Fz/PCP, Notch, and EGFR-signaling
Ommatidia are arranged with respect to the antero-posterior (A/P) and dorso-ventral (D/V) axes. While the A/P arrangement is established by progression of eye development, the D/V alignment is generated by Fz/PCP – and N and EGFR-signaling and their effectors via ommatidial rotation (OR). Preclusters emerge with a single symmetry axis, and the combination of Wnt/PCP-mediated cell fate induction and the subsequent OR realigns them at 90° from their original positio. Preclusters in dorsal and ventral halves rotate in opposite directions to create a line of mirror-symmetry across the D/V midline, the equator. Wnt/Fz-PCP mediated R3/R4 specification (defining ommatidial chirality) takes place just before OR initiates, determining direction of rotation. OR is a highly coordinated developmental cell motility process, entailing concerted regulation of force generation and associated cytoskeletal network. Its link to cell adhesion in particular E-cadherin complexes is essential. Based on genetic requirements and effects of core Fz-PCP factors in OR, i
It is assumed that the R3/R4 cells are critical in asymmetrically regulating force generation, thus driving OR directionality. Other factors, incl. N-signaling, the Nmo kinase, and the EGFR-Ras pathways are thought to “execute” these inputs and affect cell adhesion and force generation. These observations raise three significant questions, which we study in this context:
(1) What is the nature of the mechanical forces generated by the core PCP factor-effector interactions and Notch or EGFR-signaling; (2) How are these mechanically transmitted to drive a coordinated process; and (3) What are the molecular connections between the signaling pathways and junctional and cytoskeletal components during OR?
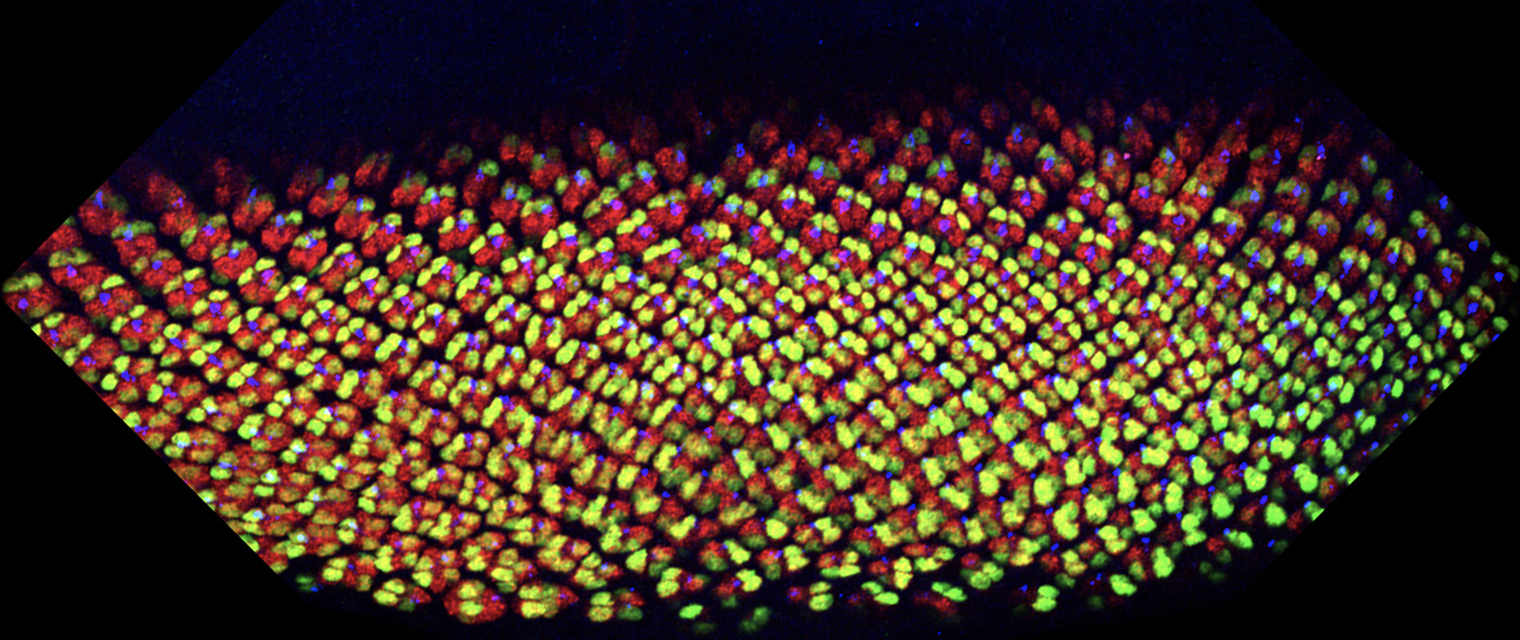
Figure: Developing eye discs stained for photoreceptor markers Elav (all in red), Svp (R3/4 and R1/6 in green) and Boss (R8). Note change in angle (rotation) of ommatdial clusters by the Svp (green) staining. Young preculsters are up and more mature ommatidia are in the lower part of picture
Featured Recent Publications
Recent Key Publications
Vuong, L., Iomini, C., Balmer, S., Esposito, D., Aaronson, S.A., and Mlodzik, M. (2018). The Kinesin -2/IFT-A complex promotes nuclear localization of b-Catenin during Wnt signaling. Nature Communications, 9, 5304. doi: 10.1038/s41467-018-07605-z. PMCID: PMC6294004
Bigenzahn, J.W, Collu, G.M., Vladimer, G.I., Heinz, L.X., Pieraks, M., Schischlik, F., Fauster, A., Rebsamen, M., Blomen, V.A., Winter, G.E. Kralovics, R., Brummelkamp, T.R., Mlodzik, M., and Superti-Furga, G. (2018). LZTR1 is a regulator of RAS ubiquitination and signaling. Science, 362, 1171-1177. PMCID in progress
Wu, J., and Mlodzik M. (2017) Wnt/PCP Instructions for Cilia in Left-Right Asymmetry. Dev Cell, 40, 423-424. PMCID: PMC5490791
Weber, U, and Mlodzik M. (2017). APC/CFzr/Cdh1–dependent Regulation of Planar Cell Polarity Establishment via Nek2 Kinase Acting on Dishevelled. Dev Cell, 40, 53-66. PMCID: PMC5225046
Carvajal-Gonzalez, J.M., Balmer, S., Mendoza, M., Dussert, A., Collu, G., Weber, U., Ciruna, B. and Mlodzik, M. (2015). The Clathrin Adapter AP-1 complex and Arf1 regulate Planar Cell Polarity in vivo. Nature Communications 6, 6751.doi: 10.1038/ncomms7751. PMCID: PMC4515971
Chin, M.L. and Mlodzik, M. (2013). Drosophila Furrowed/Selectin is a homophilic cell adhesion molecule stabilizing Frizzled and mediating its intercellular interactions during PCP establishment. Dev Cell, 26, 455-468. PMCID: PMC4084690.
Wu, J., Roman, A., Carvajal-Gonzalez, J.M., and Mlodzik, M. (2013). Wg and Wnt4 provide long-range directional input to planar cell polarity orientation in Drosophila. Nature Cell Biology, 15, 1045-1055. PMCID: PMC3762953. [with Cover]
Singh, J., Grumolato, L., Yanfeng, W.A., Aaronson, S.A., and Mlodzik, M. (2010). Abelson family kinases regulate Frizzled planar cell polarity signaling via Dsh phosphorylation. Genes & Development 24, 2157-2168. PMCID: PMC2947768 [with Cover]
Simons, M., Gault, W.J., Gotthardt, D., Rohatgi, R, Klein, T.J., Shao, Y., Lee, H.-J., Wu, A.-L., Fang, Y., Satlin, L.M., Dow, J.T., Chen, J., Zheng, J., Boutros, M., and Mlodzik, M. (2009). Electrochemical cues regulate the assembly of the Frizzled/Dishevelled complex at the plasma membrane during planar epithelial polarization. Nature Cell Biology, 11, 286-294. PMCID: PMC2803043
Meet the Team

Marek Mlodzik, PhD, Principal Investigator
Professor and Chair – Cell, Developmental & Regenerative Biology
Professor – Ophthalmology / Oncological Sciences
merek.mlodzik@mssm.edu
Contact Us
Mlodzik Laboratory
merek.mlodzik@mssm.edu
Professor, Cell, Developmental, and Regenerative Biology
Location
Annenberg 25-20
Phone
Office: 212.241.2177
Lab: 212.241.9794
