Data and Resources
SAP HANA In-Memory Database
What is SAP HANA?
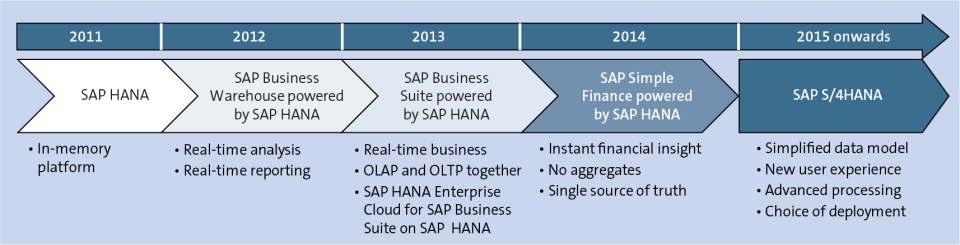
SAP HANA (High-performance Analytical Appliance) is an in-memory database platform developed by SAP SE. It uses in-memory computing to store data primarily in RAM, unlike traditional relational databases that need to retrieve data from disk-based storage solutions. With this technological innovation, SAP HANA can access in-memory data 10,000 times faster than data stored on standard disks. The result is that companies can now rapidly analyze large amounts of data and process transactions in seconds rather than hours. SAP HANA is the platform for SAP’s Enterprise Resource Planning software (S/4HANA) as well as other business applications and can run on-premises, in the cloud or in a hybrid configuration. Enterprises that have migrated over to SAP HANA have been able to realize accelerated business processes, improved data insights, and simplified IT environments. Since SAP HANA can run both OLTP and OLAP workloads, it eliminates the need to move data to another database to run analytical applications. This removes the burden of maintaining separate legacy systems, data silos and data warehouses. The result is less data redundancy, a smaller hardware footprint and less data management costs as well. SAP HANA analyzes live data for real-time business decisions and analytics, using advanced data processing engines for business, text, spatial, graph and series data. The initial version of the SAP HANA database was released to select customers in late 2010. SAP later debuted the general release at SAPPHIRE NOW, SAP’s annual technology conference in Orlando, FL in June 2011. SAP HANA made history as the first in-memory database in the world. It was an extremely popular product release, quickly becoming SAP’s fastest-adopted solution. Today there are over 30,000 customers utilizing SAP HANA.
SAP HANA Platform
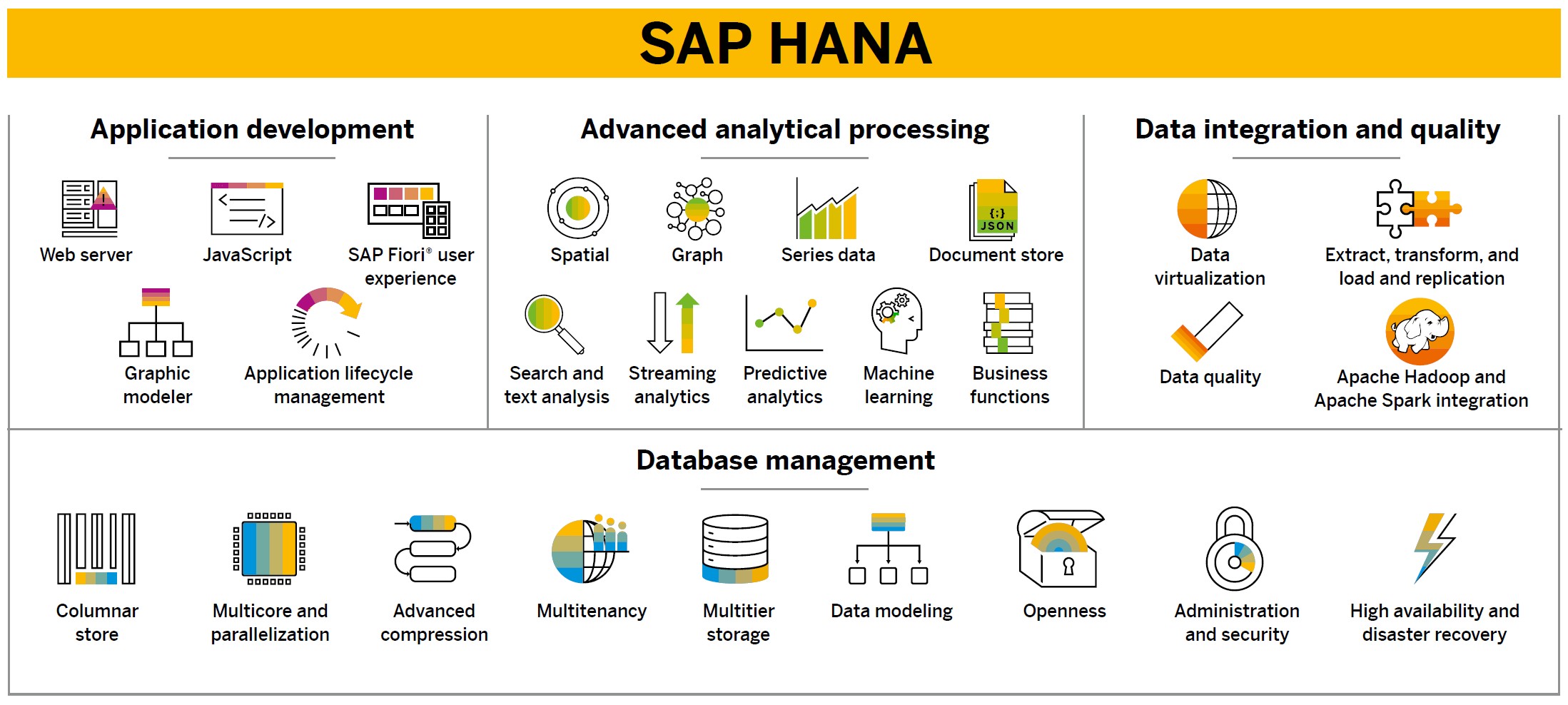
While SAP HANA is mostly recognized for its ability to process large amounts of data at record speeds, it is far more than just an in-memory database. SAP HANA offers both column and row-based storage, making it capable of handling both OLTP and OLAP workloads. Tables that are organized in columns are optimized for high performing read operations while still providing good performance for write operations. In addition, column store offers extremely efficient data compression, which in turn saves memory and speeds up searches and calculations. Typical compression rates are 7x or greater when compared with a traditional RDBMS. Other column store features include table partitioning (which can further improve performance by utilizing partition pruning), and a delta store to optimize write operations. Another technology built into the platform includes dynamic tiering, allowing for a temperature based (hot/warm/cold) data aging strategy. The purpose of this feature is to extend SAP HANA memory with a disk-centric columnar store for less frequently used data. In addition, SAP HANA offers data integration services, high availability and disaster recovery capabilities, and a complete development platform known as SAP HANA XSA (SAP HANA extended application services, advanced model) that can be used to code with Java and Node.js. It includes OData support and is the successor to the now-deprecated XS classic model. Apps that were created with SAP HANA XS can be ported over to SAP HANA XSA. The SAP Web IDE for SAP HANA is a browser-based integrated development environment used to create web-based and mobile user interfaces, business logic, and advanced data models. It provides a handful of developer tools, including syntax-aware code editors, inspection tools, debugging tools, and CDS modeling capability.
SAP HANA and Machine Learning
Python Machine Learning Client for SAP HANA
Welcome to Python machine learning client for SAP HANA (hana-ml)! This package enables Python data scientists to access SAP HANA data and build various machine learning models using the data directly in SAP HANA. This page provides an overview of hana-ml. Python machine learning client for SAP HANA consists of two main parts:
- SAP HANA DataFrame, which provides a set of methods for accessing and querying data in SAP HANA without bringing the data to the client.
- A set of machine learning APIs for developing machine learning models.
Specifically, machine learning APIs are composed of two packages:
- PAL packagePAL package consists of a set of Python algorithms and functions which provide access to machine learning capabilities in SAP HANA Predictive Analysis Library(PAL). SAP HANA PAL functions cover a variety of machine learning algorithms for training a model and then the trained model is used for scoring.
- APL packageAutomated Predictive Library (APL) package exposes the data mining capabilities of the Automated Analytics engine in SAP HANA through a set of functions. These functions develop a predictive modeling process that analysts can use to answer simple questions on their customer datasets stored in SAP HANA.
hana-ml uses SAP HANA Python driver (hdbcli) to connect to and access SAP HANA. A figure of architecture is shown below: 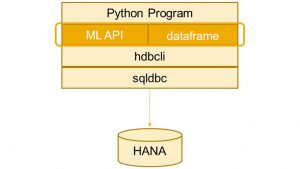
For more information, see the Python Machine Learning Client for SAP HANA website
Secure cloud infrastructure
The team at HPI.MS worked closely with engineers at Microsoft, Mount Sinai IT and Data4Life to establish secure Microsoft Azure cloud infrastructure to run AIR.MS. The following architecture diagram depicts the AIR.MS infrastructure running in the Mount Sinai Azure cloud. 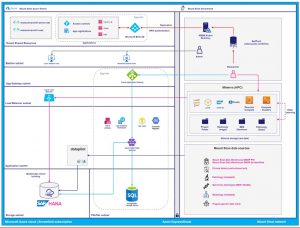
Python Libraries
SAP HANA ML (Machine Learning) is one of the Python libraries that we provide to researchers to query AIR.MS. SAP HANA ML is a component of SAP HANA, an in-memory database and analytics platform designed for handling massive amounts of data in real-time. HANA ML provides integrated machine learning capabilities to build, deploy, and execute machine learning models directly within the SAP HANA environment. It provides a wide range of machine learning algorithms, such as regression, classification, clustering, and time-series forecasting, that can be used for various use cases like patient dataset preparation, notes analysis, etc. It offers a set of libraries and APIs to develop custom machine learning models, or users can use pre-built models from the SAP HANA Model Store. The biggest advantage of HANA ML is In-Database Processing. HANA ML leverages the power of SAP HANA’s in-memory processing capabilities, allowing data to be analyzed and processed directly within the database. This eliminates the need for data movement, resulting in faster model training and predictions. HANA ML also supports model versioning and model management, allowing researchers to track and control the lifecycle of their machine learning models. Additionally, it offers model explainability features, providing insights into how the models arrive at their predictions, which is crucial for ensuring transparency and compliance with regulations. More details can be found in the SAP Documentation.
Benchmarking
AIR.MS in-memory full-text search performance examples
Number of indexed notes: 86,893,767
Example 1: Show count for notes containing an EXACT search string ‘acute kidney disease‘, broken down by gender
Example 2: Show count for notes containing FUZZY search string ‘acute kidney disease‘, broken down by gender
Example 3: Show results for FUZZY search containing term ‘ulcerative colitis‘, and group by Note Type
AIR·MS Data Modalities
AIR·MS data
The AIR·MS platform proudly features the following Mount Sinai and public datasets. Our team is dedicated to continuously expanding our database with additional data modalities, striving to build a comprehensive, multi-modal research resource. To help you get started with the data, we offer Quick Start guides available here.

Mount Sinai Data Warehouse (MSDW) in OMOP format
The MSDW dataset leverages the OMOP Common Data Model. The data is comprised of clinical data extracted from Mount Sinai’s Epic Caboodle database and other ancillary systems.
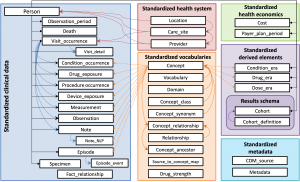
We offer both an identifiable and a de-identified version of the MSDW dataset in AIR·MS
MSDW OMOP identifiable (PHI) with extended attributes
Current Status
Schema: CDMPHI
Data snapshot from 04/16/2025
Unique patients: 12,107,621
MSDW OMOP de-identified (de-id)
Current Status
Schema: CDMDEID
Data snapshot from 09/26/2023
Unique patients: 10,934,180
| Table | Record Count |
|---|---|
| CARE_SITE | 106,699 |
| CDC_RACE_ETHNICITY_XTN | 967 |
| CONCEPT | 11,337,020 |
| CONCEPT_ANCESTOR | 101,072,304 |
| CONCEPT_CLASS | 453 |
| CONCEPT_RELATIONSHIP | 170,586,066 |
| CONCEPT_SYNONYM | 5,190,521 |
| CONDITION_OCCURRENCE | 206,228,288 |
| DEATH | 49,797 |
| DOMAIN | 50 |
| DRUG_EXPOSURE | 213,932,157 |
| DRUG_STRENGTH | 3,003,619 |
| FACT_RELATIONSHIP | 161,135,148 |
| LOCATION | 13,394,927 |
| MEASUREMENT | 1,875,881,345 |
| NOTE | 242,610,978 |
| OBSERVATION | 516,585,500 |
| OBSERVATION_PERIOD | 12,147,842 |
| PERSON | 12,107,621 |
| PROCEDURE_OCCURRENCE | 312,011,901 |
| PROVIDER | 1,363,962 |
| PROVIDER_ATTRIBUTE_XTN | 783,560 |
| RELATIONSHIP | 730 |
| VISIT_OCCURRENCE | 198,943,396 |
| VOCABULARY | 254 |
Note
Some of the standard OMOP tables contain extension fields (starting with the prefix ‘XTN’) which contain data outside of the OMOP standard data model. Many of these XTN attributes are based on data derived directly from EPIC (i.e. codes used in EPIC rather than the standardized OMOP codes), or attributes not currently contained in the OMOP standard.
| Table | Record Count |
| MEASUREMENT | 1,330,106,439 |
| OBSERVATION | 274,496,232 |
| PROCEDURE_OCCURRENCE | 192,252,154 |
| DRUG_EXPOSURE | 149,311,961 |
| CONCEPT_RELATIONSHIP | 143,179,970 |
| NOTE | 137,886,581 |
| CONDITION_OCCURRENCE | 122,966,185 |
| VISIT_OCCURRENCE | 118,936,818 |
| FACT_RELATIONSHIP | 118,710,809 |
| CONCEPT_ANCESTOR | 94,315,531 |
| CONCEPT_SYNONYM | 11,899,964 |
| OBSERVATION_PERIOD | 10,955,831 |
| PERSON | 10,934,180 |
| CONCEPT | 10,302,430 |
| DRUG_STRENGTH | 2,994,169 |
| LOCATION | 1,321,549 |
| PROVIDER | 1,307,112 |
| CARE_SITE | 301,467 |
| DEATH | 3,213 |
| RELATIONSHIP | 692 |
| CONCEPT_CLASS | 437 |
| VOCABULARY | 251 |
| DOMAIN | 50 |

Clinical Notes
Clinical notes in the form of unstructured data (progress notes, telephone encounters, nursing notes, procedures, etc.) extracted from the MSDW OMOP identifiable dataset have been loaded to AIR.MS and enabled for search using SAP HANA’s in-memory full-text search capabilities. This feature empowers the researcher to build patient cohorts based on terms contained in unstructured reports in seconds or even milliseconds! The researcher can further filter based on note type or an array of other clinical attributes.
Tip: See the Getting Started Guides for examples on how to perform search using python!
Current Status
Schema: CDMPHI
Data snapshot from 04/16/2025
Number of indexed notes: 242,610,978

Pathology (metadata)
The Pathology metadata aids researchers in the field of Computational Pathology. Researchers are able to query the metadata in combination with other linked data modalities to build a patient cohort, subsequently apply quantitative methods for the analysis of digital microscopy slides and relating the resulting statistical descriptors to patient outcomes.
We are also working on making the digital slides available to researchers on Minerva HPC.
Current Status
Data Source: Powerpath
Schema: CDMPATHOLOGY
Data snapshot from 01/08/2025
Unique patients: 3,528,719
Note
Pathology reports are now available in AIR·MS and can be found in table ACC_RESULTS. The reports are broken up into sections (clinical history, final diagnosis, SNOMED coding, etc.) that can be identified by column PATH_RPT_HEADING_NAME. If you are looking for the full report, you can combine all records for with the same ACCESSION_2_ID into a single output. The column containing the free-text (ACC_RESULTS_FINDING) has been enabled with SAP HANA full-text search capabilities. Examples of how to use HANA full-text search are available in this tutorial notebook. The following tables and attributes are available in AIR·MS: ACCESSION
Number of records: 7,107,464
| Column Name | Comments |
|---|---|
| ACC_CATG | Case category |
| ACC_PROCESS_STEP_COMPLETED_DATE | Case finalize date / status update datetime |
| ACCESSION_2_ID | PowerPath unique case ID |
| ACCESSION_NO | Case number |
| BIRTH_DATE | Patient date of birth |
| CREATED_DATE | Case creation date |
| CURRENT_STATUS_ID | PowerPath unique identifier for a case status |
| FACILITY_CODE | Facility code |
| FACILITY_ID | PowerPath ID for the facility associated with the accession |
| FACILITY_NAME | Facility name |
| IMPORTED_CASE | One-character \”Y\” or \”N\” code indicating if the case was imported into PowerPath |
| LAST_UPDATE_DATETIME | Case finalize date / status update datetime |
| MED_REC_NO | Medical record number (EPIC MRN) |
| MRN_FACILITY_CODE | PowerPath ID for the facility that assigned the MRN |
| MRN_FACILITY_DESCRIPTION | Name of the facility that assigned the MRN |
| ORDER_NUMBER | Order number from the ordering system |
| PATIENT_AGE | The patient’s age on the case creation date |
| PATIENT_ID | PowerPath patient ID |
| PERSONNEL_2_FULL_NAME | Name of the pathologist who finalized the accession |
| PERSONNEL_2_ID | PowerPath ID for the pathologist who finalized the accession |
| PROCESS_STEP_DESCRIPTION | Case status name / description |
| VISIT_NUMBER | Encounter identifier |
ACC_ICD
Number of records: 3,548,815
| Column Name | Comments |
|---|---|
| ACC_ICD9_ID | PowerPath surrogate unique identifier for an ICD-10 code assigned to a case |
| ACCESSION_2_ID | PowerPath unique case ID |
| LAST_UPDATE_DATETIME | Case finalize date / status update datetime |
| MEDICAL_CODE | ICD-10 code assigned for billing |
| MEDICAL_CODE_ID | PowerPath surrogate unique identifier for an ICD-10 code |
ACC_SLIDE
Number of records: 14,241,494
| Column Name | Comments |
|---|---|
| ACC_BLOCK_ID | PowerPath unique block ID |
| ACC_BLOCK_LABEL | Specimen block identifier |
| ACC_PROCESS_STEP_COMPLETED_DATE | Case finalize date / status update datetime |
| ACC_SLIDE_ID | PowerPath unique slide ID |
| ACC_SPECIMEN_DESCRIPTION | Specimen source description |
| ACC_SPECIMEN_ID | PowerPath unique specimen ID |
| ACCESSION_2_ID | PowerPath unique case ID |
| BIOPSY | Boolean flag for 1 = biopsy, 0 = non-biopsy |
| COLLECTION_DATE | Specimen collection date |
| CONSULT_LABEL | Optional free text for slides of type \”consult\” |
| LAB_PROCEDURE_CODE | The procedure code |
| LAB_PROCEDURE_DESCRIPTION | The procedure description |
| LAB_PROCEDURE_ID | PowerPath unique procedure identifier |
| LAST_UPDATE_DATETIME | Case finalize date / status update datetime |
| RECV_DATE | Specimen received date |
| SLIDE_LABEL | Derived unique (business key) identifier for each slide |
| SLIDE_NO | Ordinal number of the slide from the specimen & block |
| SLIDE_TYPE | Whether the slide is stained, unstained or antibody/IHC |
| SOURCE_MATERIAL_LABEL | Derived unique (business key) identifier for each slide’s source specimen and block |
| SOURCE_REC_TYPE | Where the slide came from, either specimen or block |
| SPECIMEN_CATEGORY_ID | PowerPath specimen category ID |
| SPECIMEN_CATEGORY_NAME | The specimen category name |
| SPECIMEN_GROUPS_CODE | Specimen specialty code |
| SPECIMEN_GROUPS_ID | PowerPath specimen specialty ID |
| SPECIMEN_LABEL | Specimen identifier |
| TYPE | Whether the slide is consult or not consult |
ACC_RESULTS
Number of records: 31,193,765
| Column Name | Comments |
|---|---|
| ACC_RESULTS_FINDING | The text of the report section |
| ACC_RESULTS_ID | PowerPath unique identifier for each report section |
| ACC_RESULTS_REC_ID | sort order of result section on RTF |
| ACCESSION_2_ID | PowerPath unique case ID |
| LAST_UPDATE_DATETIME | Case finalize date / status update datetime |
| PATH_RPT_HEADING_ID | PowerPath result section heading on RTF |
| PATH_RPT_HEADING_NAME | PowerPath result section heading name |
ACC_SLIDE_IMAGESERVER
Number of records: 2,564,239
| Column Name | Comments |
|---|---|
| ACC_SLIDE_ID | PowerPath unique slide ID |
| ACC_SLIDE_IMAGESERVER_DESCRIPTION | The name of the Philips iSyntax slide image file |
| ACC_SLIDE_IMAGESERVER_ID | PowerPath unique identifier for a slide image |
| INTERNAL_SLIDE_ID | Identifier for the slide, also known as the \”barcode\” ID |
| LAST_UPDATE_DATETIME | Case finalize date / status update datetime |
| SCAN_DATE | The date on which the slide image was digitized |

Radiology (metadata)
AIR·MS now features radiology metadata extracted from the Mount Sinai IRW 2.0 XNAT system (via an MSDW data pipeline). This data set is comprised of detailed DICOM (Digital Imaging and Communications in Medicine) tags associated with the medical images.
These tags provide essential metadata, including patient information, imaging parameters, equipment details, and procedural context, ensuring a comprehensive understanding of each radiological study.
By integrating this metadata, we enable researchers to gain deeper insights into the imaging data, facilitating advanced analyses and fostering innovations in medical imaging research.
Current Status
Schema: CDMRADIOLOGY
Data snapshot from 8/16/2024
Unique patients: 2,057,482
The following tables and attributes are available in AIR·MS:
Number of records: 66,993,826
| RADIOLOGY_METADATA |
| ID |
| PATIENT_ID |
| SERIES_INSTANCE_UID |
| STUDY_INSTANCE_UID |
| ETL_RECORD_UPDATE_DATETIME |
Number of records: 7,745,407,087
| RADIOLOGY_DICOM_DATA |
| ID |
| RADIOLOGY_METADATA_ID |
| DICOM_TAGS |
| TAG_VALUE_REPRESENTATION |
| TAG_VALUE” NCLOB MEMORY |
| TAG_XTN_PATIENT_EPIC_MRN |
| TAG_PATIENT_NAME |
| SERIES_INSTANCE_UID |
| STUDY_INSTANCE_UID |
| ETL_RECORD_UPDATE_DATETIME |
Mount Sinai Million Health Discoveries Program
The current lack of diversity in genomic research data is hindering what we can learn about health and potential treatments in our global population. By enhancing the diversity of people participating in genomic research, we can advance our knowledge and discovery of human genetics for all populations. To that end, The Charles Bronfman Institute for Personalized Medicine is spearheading the effort to carry out the genetic sequencing of one million Mount Sinai patients within the next five years. This initiative, one of the largest such sequencing projects of its kind, will integrate health and research data at Mount Sinai to promote discoveries that will directly benefit our patient population.
Access to the BioMe Biobank and Mount Sinai Million Biobank on HPC can be requested via the CBIPM Data and Specimen Inquiry Form
Mount Sinai Million / BioMe identifiable (PHI)
Current Status
Schema: CDMMSM
Data snapshot from 06/09/2025
Unique patients: 279,885
Mount Sinai Million / BioMe de-identified (de-id)
Current Status
Schema: CDMMSMDEID
Data snapshot from 06/09/2025
Unique patients: 279,885
| PATIENT | Comments |
| ID | Internal ID for linking to other tables within the dataset |
| MRN | Medical Record Number (EPIC MRN – only accessible under regulatory approval) |
| MASKED_MRN | De-Identifier for combined BioMe Biobank set with Regeneron and Sema4 data |
| RGN_ID | De-identifier for first Regeneron batch regarding BioMe Biobank |
| SEMA4_ID | De-identifier for Sema4, a subset of Masked MRN ID |
| MSM_ID | De-identifier for Mount Sinai Million Biobank, a combined setoff RGN_ID and new MSM ID |
| MILLION_ID | Indicator for all consented patients with and without genomic data |
| AIR_CREATED_AT | Record creation in AIR·MS |
| AIR_UPDATED_AT | Record updated in AIR·MS |
| PATIENT | Comments |
| ID | Internal ID for linking to other tables within the dataset |
| MASKED_MRN | De-Identifier for combined BioMe Biobank set with Regeneron and Sema4 data |
| RGN_ID | De-identifier for first Regeneron batch regarding BioMe Biobank |
| SEMA4_ID | De-identifier for Sema4, a subset of Masked MRN ID |
| MSM_ID | De-identifier for Mount Sinai Million Biobank, a combined setoff RGN_ID and new MSM ID |
| MILLION_ID | Indicator for all consented patients with and without genomic data |
| AIR_CREATED_AT | Record creation in AIR·MS |
| AIR_UPDATED_AT | Record updated in AIR·MS |

Electrocardiogram (ECG)
Data Source: GE HealthCare MUSE Cardiology Information System
Schema: CDMECG
Data snapshot from: 04/10/2021
Unique patients: 1,961,254
The following tables and attributes are available in AIR·MS:
Number of records: 9,275,130
| PATIENT_DEMOGRAPHICS |
| PATIENT_DEMOGRAPHICS_ID (X) |
| FILE_ENTRY_ID |
| PATIENT_ID |
| PATIENTAGE |
| AGEUNITS |
| DATEOFBIRTH |
| GENDER |
| RACE |
| PATIENTLASTNAME |
| PATIENTFIRSTNAME |
Number of records: 9,168,266
| DIAGNOSIS |
| DIAGNOSIS_ID (X) |
| FILE_ENTRY_ID |
| MODALITY |
| DIAGNOSISSTATEMENT |
Number of records: 73,631,055
| LEAD_DATA |
| LEAD_DATA_ID (X) |
| FILE_ENTRY_ID |
| LEADBYTECOUNTTOTAL |
| LEADTIMEOFFSET |
| LEADSAMPLECOUNTTOTAL |
| LEADAMPLITUDEUNITSPERBIT |
| LEADAMPLITUDEUNITS |
| LEADHIGHLIMIT |
| LEADLOWLIMIT |
| LEADID |
| LEADOFFSETFIRSTSAMPLE |
| FIRSTSAMPLEBASELINE |
| LEADSAMPLESIZE |
| LEADOFF |
| BASELINESWAY |
| LEADDATACRC32 |
| WAVEFORMDATA |
Number of records: 9,610,935
| ECG_FILES |
| FILE_ENTRY_ID (X) |
| FILE_NAME |
| FILE_PATH |
| FILE_HASH |
| FILE_SIZE_BYTES |
| ACQUISITION_DATE |
| ACQUISITION_TIME |
| PROCESSING_STATUS |
| STATUS_CODE |
| NOTES_AND_COMMENTS |
| FILE_TIMESTAMP |
| AIR_CREATED_AT |
| AIR_UPDATED_AT |
| JSON_STATUS |
Number of records: 9,168,230
| MUSE_INFO |
| MUSEVERSION |
| FILE_ENTRY_ID |
Number of records: 7,757,472
| ORDER_INFO |
| ORDER_INFO_ID (X) |
| FILE_ENTRY_ID |
| HISACCOUNTNUMBER |
| ORDERTIME |
| ADMITTIME |
| ADMITDATE |
| HISLOCATION |
| BED |
| ATTENDINGMDHISID |
| ATTENDINGMDLASTNAME |
| ATTENDINGMDFIRSTNAME |
| ALTERNATEVISITID |
| HISDISPOSITION |
| ADMITSOURCE |
| PRIMARYDIAGNOSTICCODE |
| SERVICINGFACILITY |
| ADMITTINGMDHISID |
| ADMITTINGMDLASTNAME |
| ADMITTINGMDFIRSTNAME |
| CONSULTINGMDID |
| REFERRINGMDHISID |
| HOSPITALSERVICE |
| ADMISSIONTYPE |
Number of records: 9,164,220
| ORIGINAL_DIAGNOSIS |
| ORIGINAL_DIAGNOSIS_ID (X) |
| FILE_ENTRY_ID |
| MODALITY |
| DIAGNOSISSTATEMENT |
Number of records: 9,168,044
| ORIGINAL_RESTING_ECG_MEASUREMENTS |
| ORIGINAL_RESTING_ECG_MEASUREMENTS_ID (X) |
| VENTRICULARRATE |
| ATRIALRATE |
| PRINTERVAL |
| QRSDURATION |
| QTINTERVAL |
| QTCORRECTED |
| PAXIS |
| RAXIS |
| TAXIS |
| QRSCOUNT |
| QONSET |
| QOFFSET |
| PONSET |
| POFFSET |
| TOFFSET |
| ECGSAMPLEBASE |
| ECGSAMPLEEXPONENT |
| QTCFREDERICA |
Number of records: 3,613,372
| PHARMA_DATA |
| PHARMA_DATA_ID (X) |
| PHARMARRINTERVAL |
| PHARMAUNIQUEECGID |
| PHARMAPPINTERVAL |
| PHARMACARTID |
| FILE_ENTRY_ID |
Number of records: 9,649,712
| QRS_TIMES_TYPES |
| GLOBALRR |
| QTRGGR |
| FILE_ENTRY_ID |
Number of records: 9,657,368
| RESTING_ECG |
| RESTING_ECG_ID (X) |
| FILE_ENTRY_ID |
| PATIENT_ID |
| ACQUISITIONDATE |
| ACQUISITIONTIME |
| STATUS |
Number of records: 9,167,778
| RESTING_ECG_MEASUREMENTS |
| RESTING_ECG_MEASUREMENTS_ID (X) |
| FILE_ENTRY_ID |
| VENTRICULARRATE |
| ATRIALRATE |
| PRINTERVAL |
| QRSDURATION |
| QTINTERVAL |
| QTCORRECTED |
| PAXIS |
| RAXIS |
| TAXIS |
| QRSCOUNT |
| QONSET |
| QOFFSET |
| PONSET |
| POFFSET |
| TOFFSET |
| ECGSAMPLEBASE |
| ECGSAMPLEEXPONENT |
| QTCFREDERICA |
Number of records: 9,654,325
| TEST_DEMOGRAPHICS |
| TEST_DEMOGRAPHICS_ID (X) |
| FILE_ENTRY_ID |
| DATATYPE |
| SITE |
| SITENAME |
| ACQUISITIONDEVICE |
| STATUS |
| EDITLISTSTATUS |
| PRIORITY |
| LOCATION |
| LOCATIONNAME |
| ROOMID |
| ACQUISITIONTIME |
| ACQUISITIONDATE |
| CARTNUMBER |
| ACQUISITIONSOFTWAREVERSION |
| ANALYSISSOFTWAREVERSION |
| EDITTIME |
| EDITDATE |
| EDITORID |
| REFERRINGMDLASTNAME |
| REFERRINGMDFIRSTNAME |
| ACQUISITIONTECHLASTNAME |
| EDITORLASTNAME |
| EDITORFIRSTNAME |
| SECONDARYID |
| HISSTATUS |
/482268916-Echocardiogram-image-scaled.jpeg)
Echocardiogram (echo)
Schema: CDMECHO
Data snapshot from: 10/17/2023
Unique patients: 885,957
The following tables and attributes are available in AIR·MS:
Number of records: 268,682,931
| ECHO_METADATA |
| ID |
| FILE_ENTRY_ID |
| PATIENT_ID |
| SERIES_INSTANCE_UID |
| STUDY_INSTANCE_UID |
| SOP_INSTANCE_UID |
| IMAGE_FILE_PATH |
| AIR_CREATED_AT |
| AIR_UPDATED_AT |
Number of records: 21,089,507,788
| ECHO_TAGS_DATA |
| ID |
| FILE_ENTRY_ID |
| SERIES_INSTANCE_UID |
| STUDY_INSTANCE_UID |
| SOP_INSTANCE_UID |
| TAGS |
| TAG_VALUE |
| AIR_CREATED_AT |
| AIR_UPDATED_AT |
/3616198284.png)
Synthetic Public Use File (DE-SynPUF)
The SYNPUF (Synthetic Public Use Files) dataset, provided by the Centers for Medicare & Medicaid Services (CMS), offers a synthetic version of Medicare claims data from the years 2008 to 2010. This dataset is meticulously designed to maintain the statistical properties and relationships present in the original data while ensuring that no actual patient information is disclosed, thereby safeguarding privacy.
SYNPUF includes a comprehensive array of variables such as beneficiary demographics, chronic conditions, hospital and outpatient claims, and prescription drug events, making it an invaluable resource for researchers and data scientists. It serves as an exemplary tool for developing and testing healthcare models, algorithms, and applications without the constraints associated with sensitive real-world data.
The SYNPUF dataset in AIR·MS utilizes the OMOP Common Data Model, aligned with other clinical data sets available on the platform. Since SYNPUF data does not require an approved IRB, you can easily get onboarded and start building ML models!
Current Status
Schema: CDMSYNPUF
Number of patients: 2,326,856
Number of observations: 37,531,051
Number of measurements: 72,387,791

Work in Progress
We are constantly integrating new data to AIR·MS. The following data modalities are currently being worked on:
-
Electroencephalogram (EEG)
-
Endoscopy & colonoscopy reports
-
Bedmaster
-
Radiology images and reports





