 Kidney diseases manifest in progressive loss of renal function, which ultimately leads to complete kidney failure. Multiple factors involved in the pathogenesis of kidney diseases have made the traditional candidate gene approach of limited value toward full understanding of the molecular mechanisms of these diseases. A systems biology approach that integrates computational modeling with large-scale data gathering of the molecular changes is useful in identifying the multiple interacting genes and their products that drive kidney diseases.
Kidney diseases manifest in progressive loss of renal function, which ultimately leads to complete kidney failure. Multiple factors involved in the pathogenesis of kidney diseases have made the traditional candidate gene approach of limited value toward full understanding of the molecular mechanisms of these diseases. A systems biology approach that integrates computational modeling with large-scale data gathering of the molecular changes is useful in identifying the multiple interacting genes and their products that drive kidney diseases.
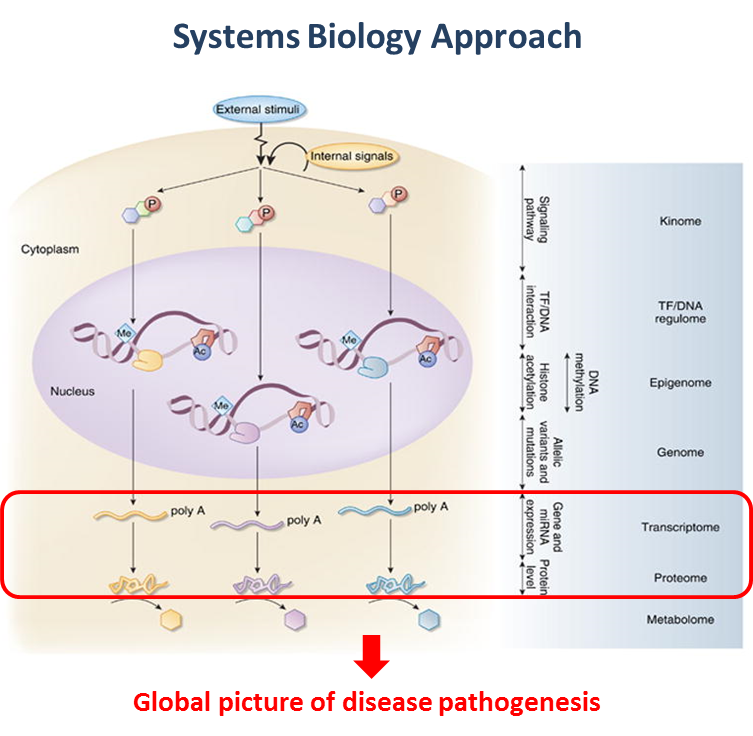
We have been using systems biology approach to study cell signaling and transcription networks in kidney cells. We have performed transcriptomic and proteomic studies of isolated glomeruli and sorted glomerular cells from animal models of DKD in order to identify key molecules involved in the pathogenesis of kidney disease. We developed an innovative program to deduce upstream signaling network from transcriptomic datasets. Using this approach, we identified HIPK2 as a potential drug target for renal fibrosis. We also develop the computational program to predict drug combination therapy in animal models of kidney disease. In collaboration with Dr. Barbara Murphy, we found that a CK- associated intronic locus in SHROOM3 gene contributes to renal fibrosis through activation of TCF7-mediated pathway. Using the systems biology approach, we also identified RTN1 as a key molecule involved in ER stress and apoptosis of kidney cells in DKD.
