
Our Research
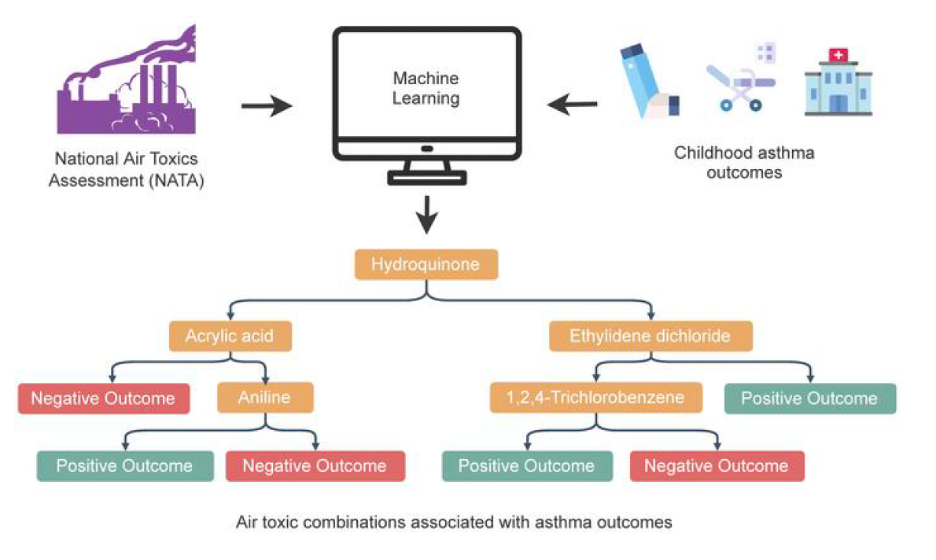
We identified combinations of early-life air pollutants associated with poor asthma outcomes in later childhood by applying a novel machine-learning method called Data-driven ExposurE Profile extraction (DEEP). Read more about it here in the Journal of Clinical Investigation.
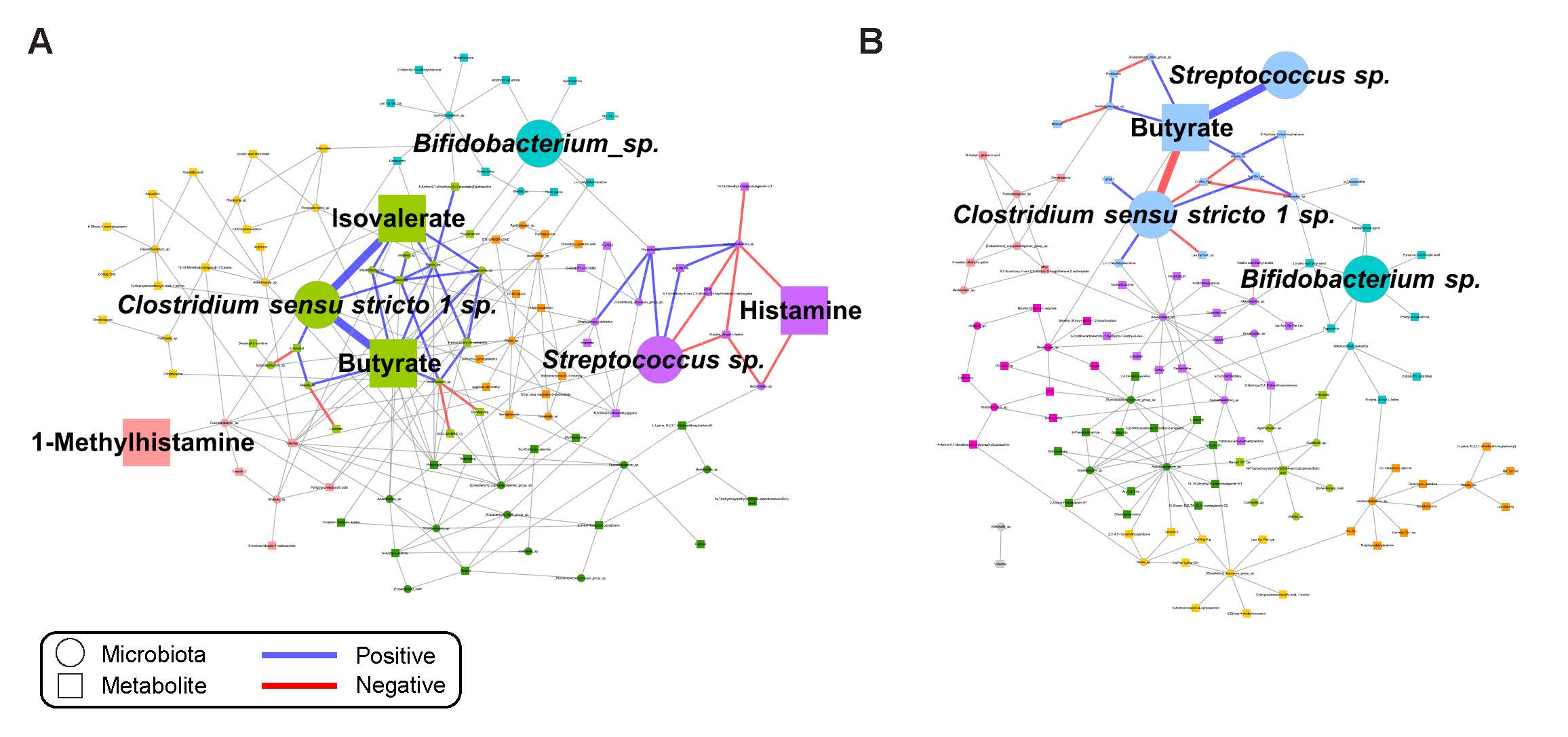
Our multi-center, longitudinal study of children from infancy through mid-childhood revealed distinct trajectories and relationships between the gut microbiome and gut metabolome in children who did and did not develop peanut allergy. Click here for the full text.
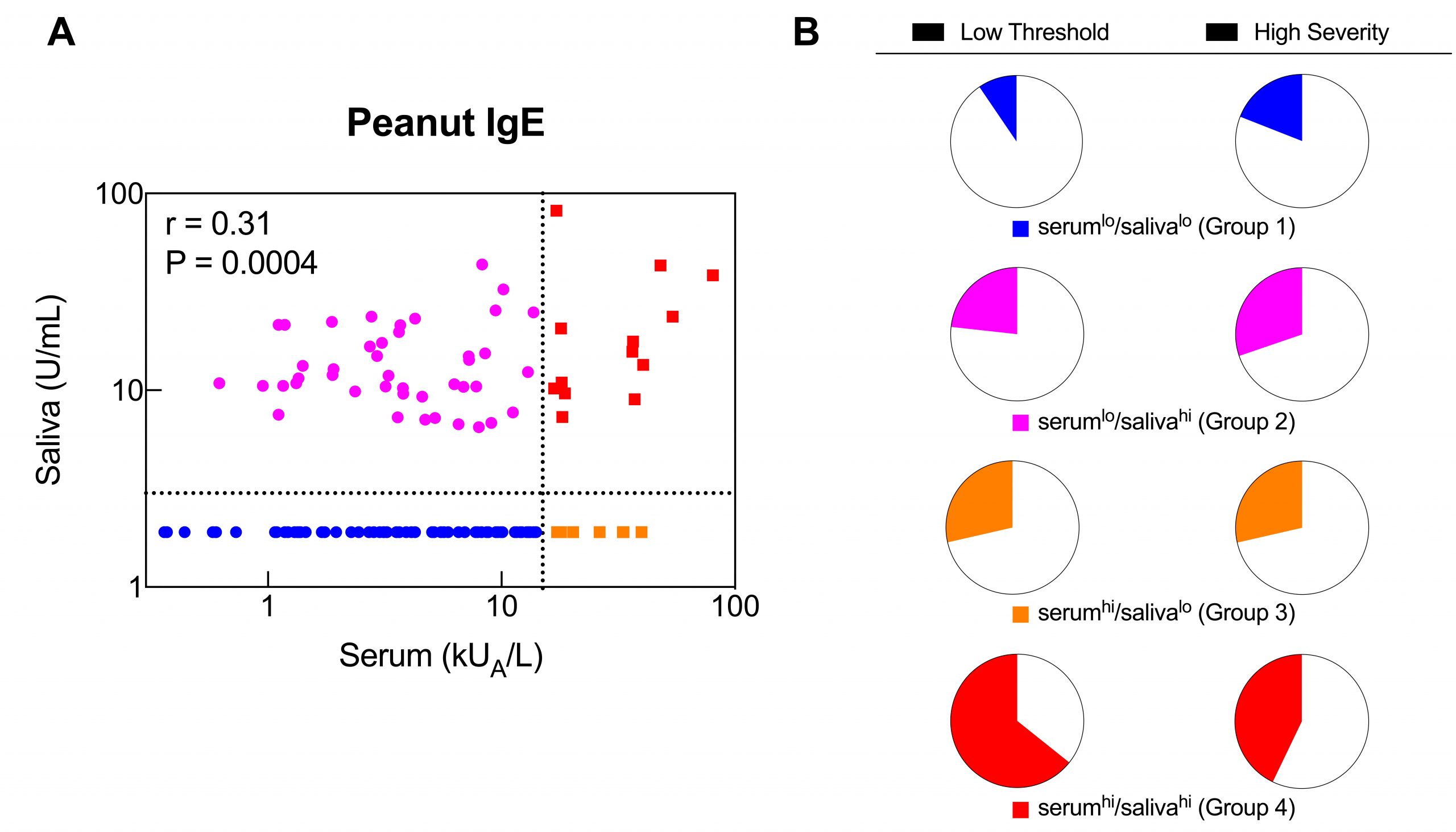
We found that peanut-specific antibody levels measured in easy-to-collect saliva are associated with the severity and threshold of reactions in peanut allergic children. These findings could inform non-invasive biomarker development for food allergy. More about the study here.
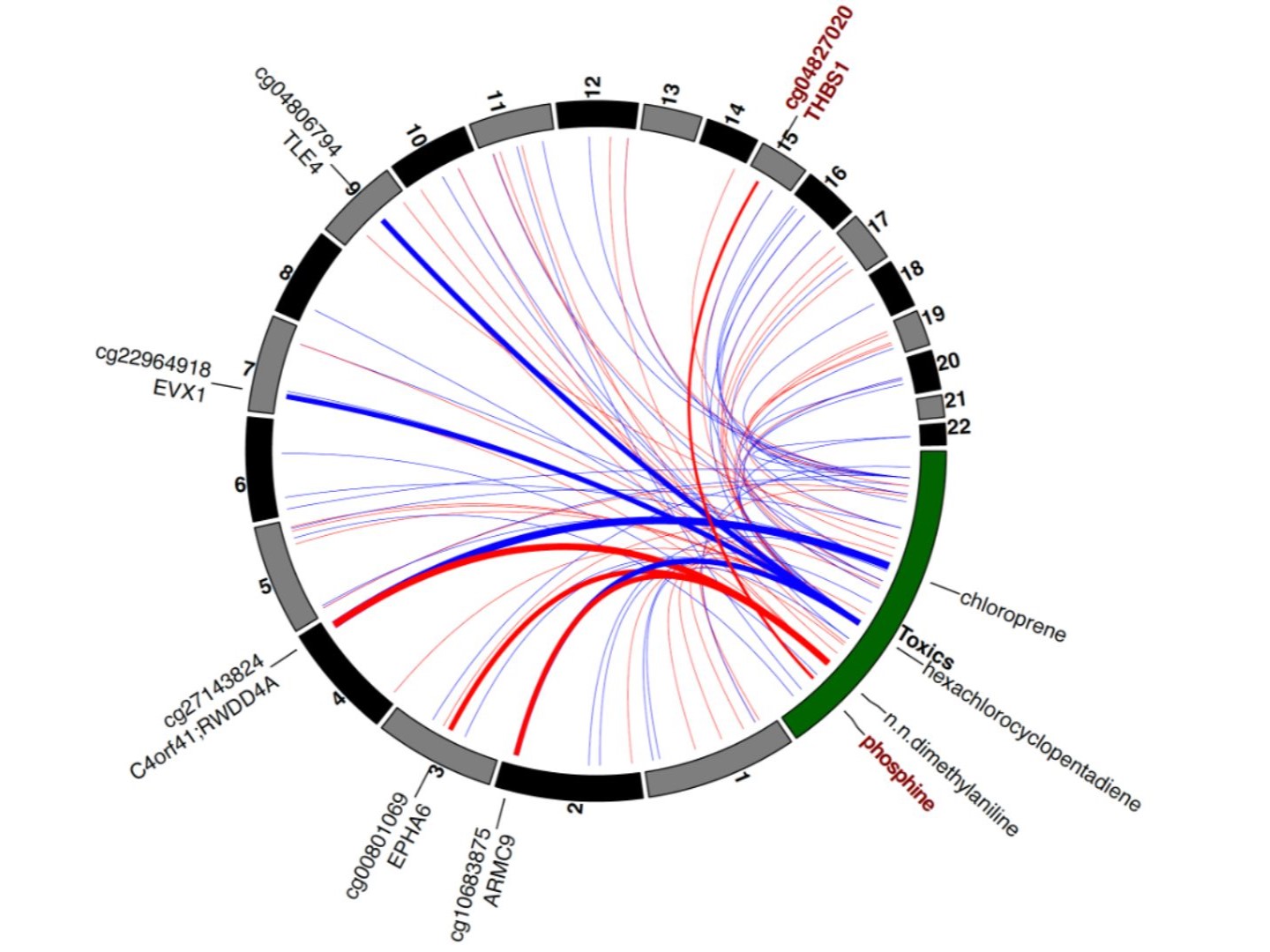
Our multi-omic integrative analysis of early-life air toxic exposures, airway methylome, airway transcriptome, and allergic rhinitis identified novel alterations and causal relationships in nasal mucosal biology that mediate air pollutant effects on allergic rhinitis. Read more about it here.
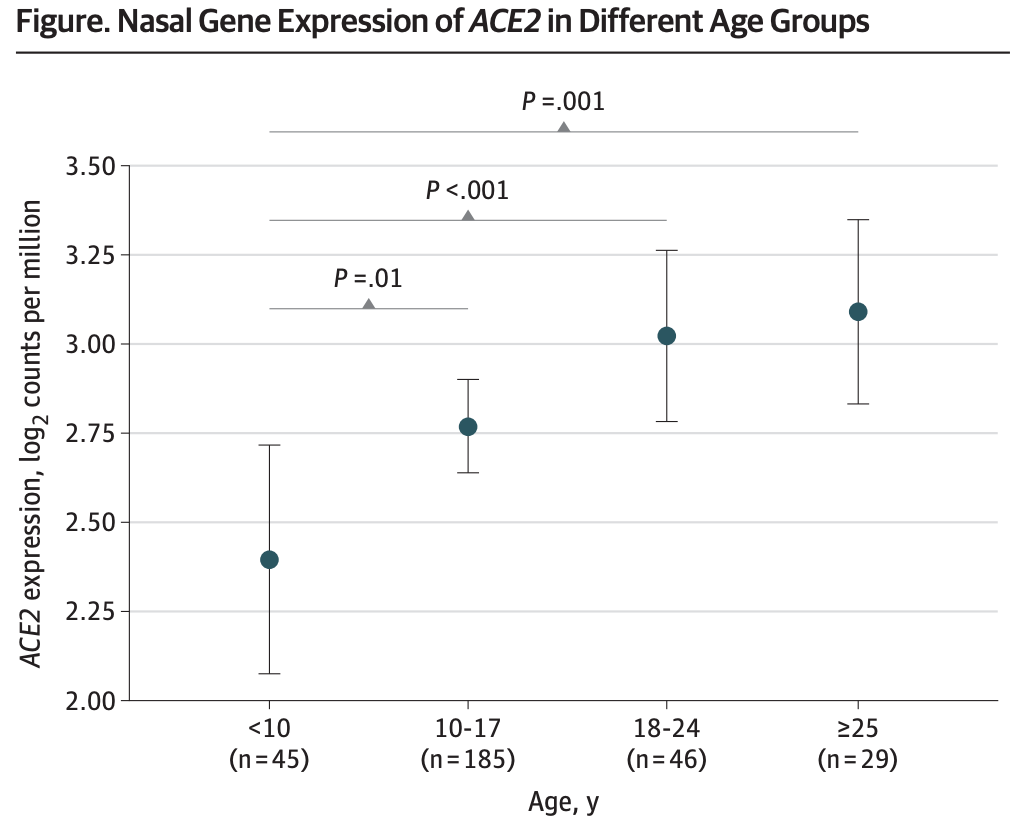
In this publication in JAMA, we report that nasal gene expression of ACE2, the receptor that the coronavirus SARS-CoV-2 uses to enter the host, is lowest in young children and increases with age. This may partially explain why children have suffered less from COVID-19. Clear here for the full text.
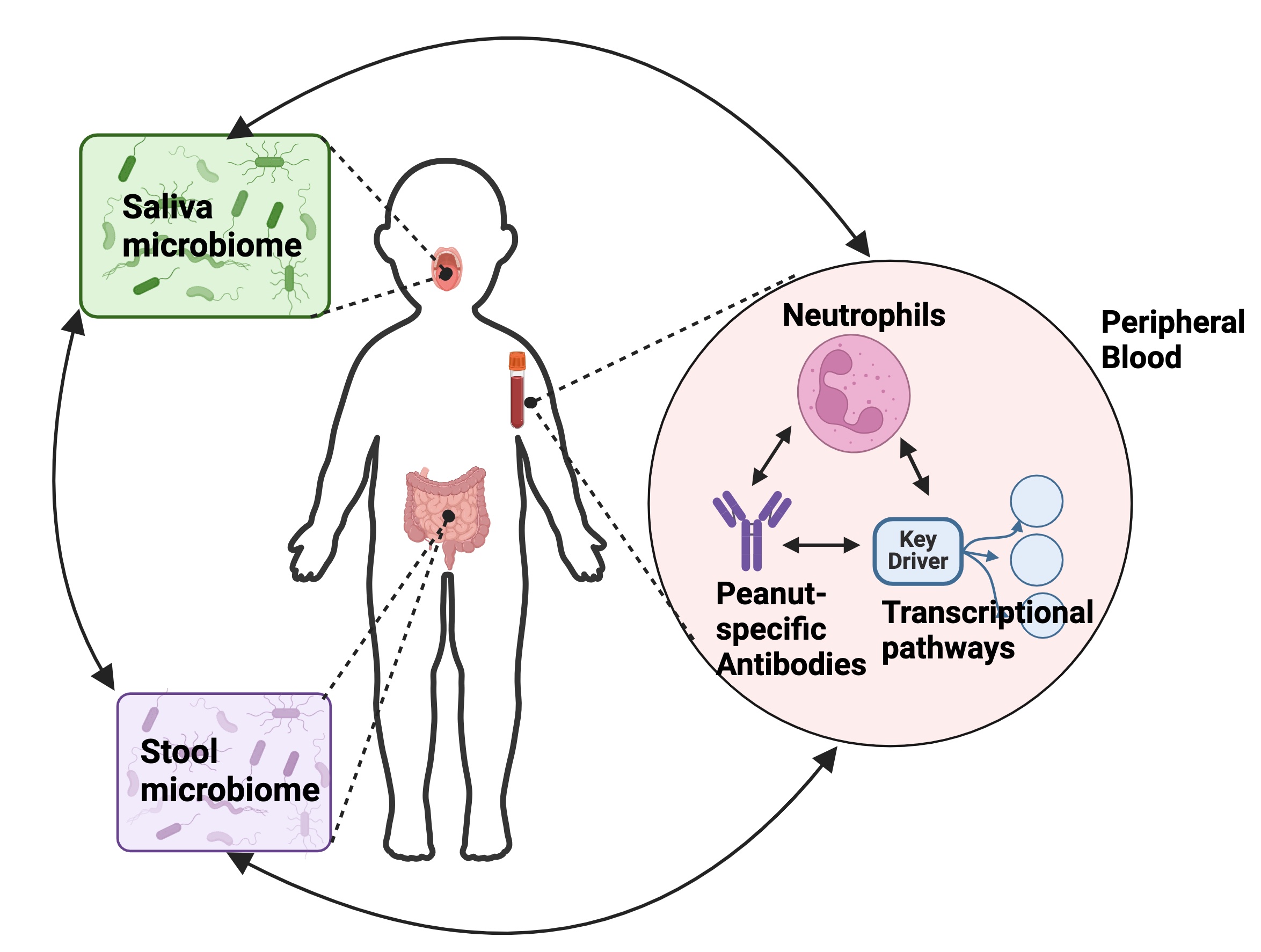
This systems biology study of oral and stool microbiomes, blood transcriptome, cellular profiles, and peanut-specific serum antibodies elucidated new relationships between local microbiota and systemic measures associated with reaction threshold in peanut allergy. Read more about it here.
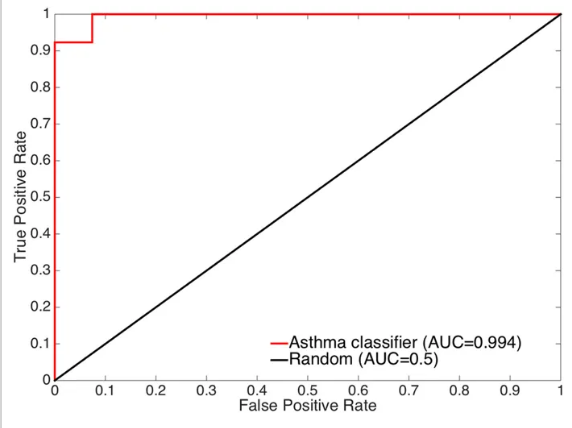
Using machine learning and nasal RNA sequence data from our well-characterized asthma cohorts, we identified and tested a nasal brush-based classifier of asthma. Click here to read the article.
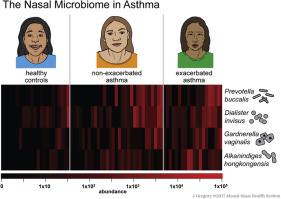
Our research group has characterized how the nasal microbiome varies with asthma activity. Read more here.
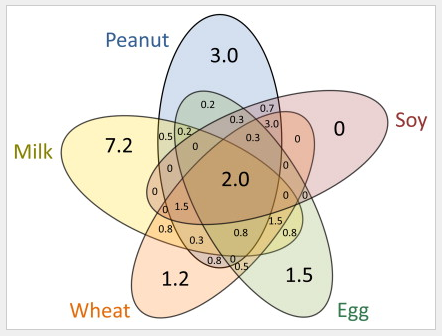
Our examination of maternal dietary habits during pregnancy in over 1000 mother-child pairs supports that pregnant women should not avoid specific foods during pregnancy to reduce the risk of their children developing asthma and allergies. Click here for the full text.
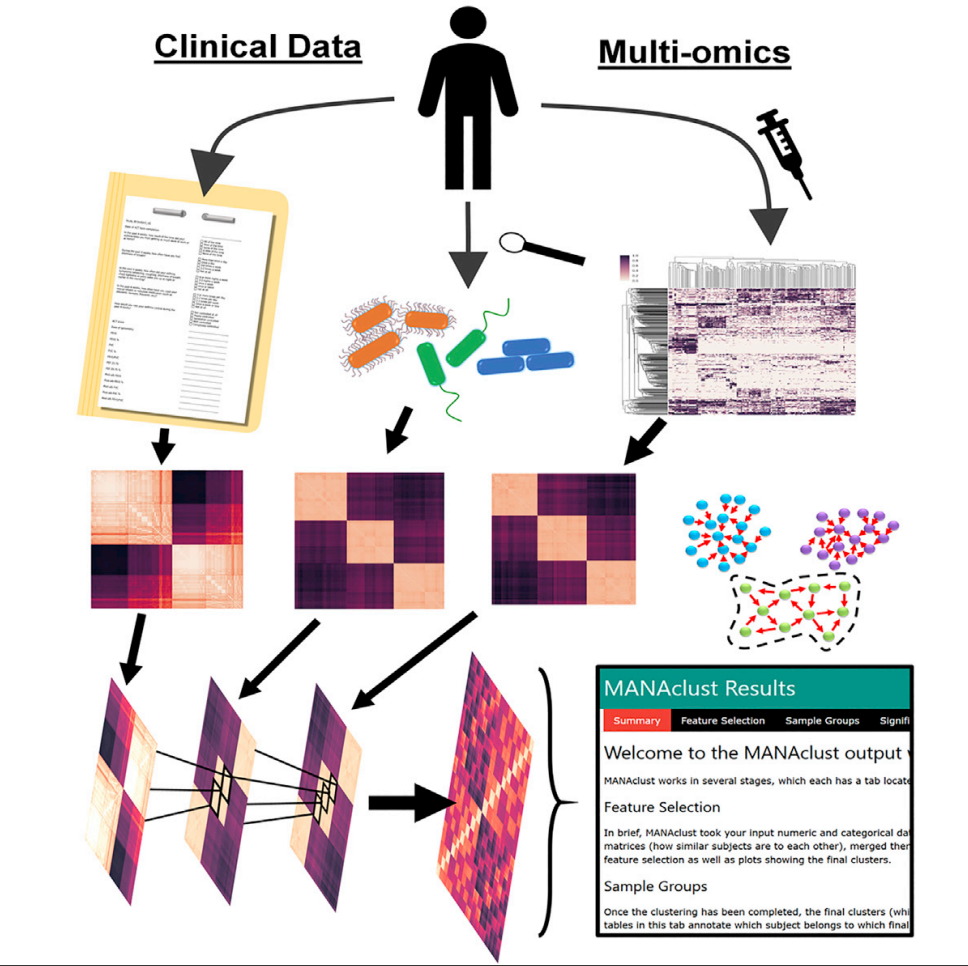
Merged Affinity Network Association Clustering (MANAclust) is a coding-free, automated pipeline developed by us that enables integration of multi-omic and clinical data clustering to identify disease endotypes. Full article in Cell Reports here.
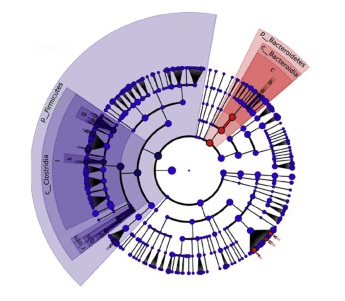
Our examination of early-life gut microbiota in milk allergic children from a longitudinal, multicenter observational study showed that Clostridia and Firmicutes are associated with resolution of milk allergy. Click here for more information.
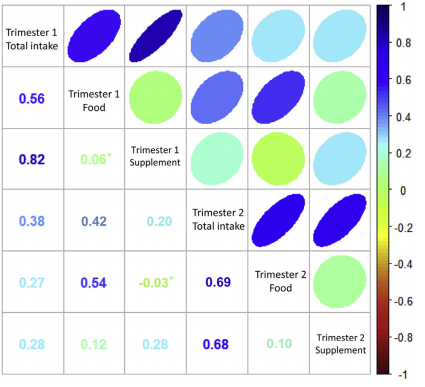
We found that food-based vitamin D — but not supplemental vitamin D– in maternal diets during pregnancy was associated with lower rates of hay fever in children at school age. Read more here.
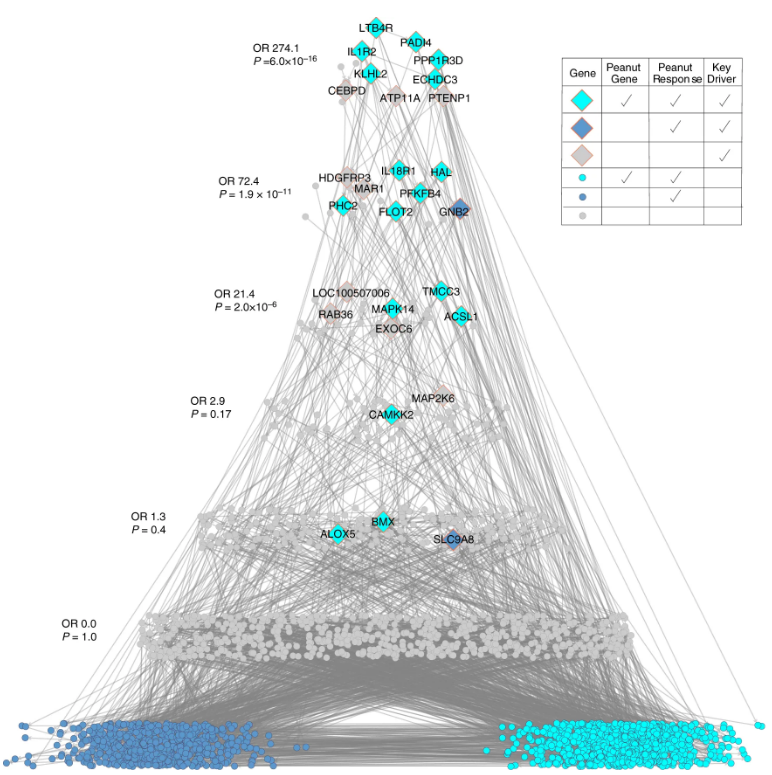
We constructed causal networks using peripheral blood transcriptomes of peanut allergic children undergoing randomized, double-blind, placebo-controlled oral challenges to detect key drivers and putative therapeutic targets of peanut allergic reactions. Read more here.
