Laboratory of A-to-I Editing and Rare Disorders
Our research is centered on unraveling the complexities of RNA editing and the genetics/genomics underlying brain aging and neurological disorders, with a strong emphasis on rare neurodevelopmental disorders. We employ an integrative approach, drawing from human neurogenetics, computational biology, molecular biology, and cellular neuroscience.
Research Focus Areas:
1. We conduct cutting-edge genomic studies to uncover the mechanisms of monogenic neurodevelopmental disorders.
2. Our team develops oligonucleotide-directed RNA editing therapeutics to address the molecular underpinnings of these disorders.
3. We leverage large-scale transcriptomic technologies to dissect the function and regulation of RNA editing in brain development, disease, and aging.
Lab Culture and Philosophy:
In our lab, we are committed to fostering a positive, inclusive, and collaborative lab environment that prioritizes professional growth and work/life balance. As a mentor, I am dedicated to training and inspiring the next generation of scientists, whether they pursue careers in academia or industry – everyone’s journey is different! Ultimately, we aim to enable all lab members to make meaningful scientific contributions while engaging in rigorous, ethical, and innovative research. Our lab thrives as part of a highly collaborative ecosystem, working alongside computational and experimental biologists within the Seaver Autism Center for Research and Treatment, the MINDICH Child Health and Development Institute, and the Departments of Genetics and Genomic Sciences and Psychiatry at Mount Sinai.
Research Topics:
- RNA biology
- Adenosine-to-Inosine (A-to-I) editing and ADAR
- Alternative splicing, gene expression, protein function
- Antisense oligonucleotides (ASOs)
- Brain development, brain aging, brain/neurological disorders
- Autism spectrum disorder and syndromic subtypes
- Rare disorders
- Neurodegeneration
- Immunogenetics of developmental disorders
- Quantitative trait loci
Genomic Technologies and Methods:
Our lab utilizes a diverse array of cutting-edge technologies and methodologies, including:
____________________________________________________
Michael S. Breen, PhD (he/him)
Associate Professor
I am the Principal Investigator of a multidisciplinary team at the forefront of human genetics, genomics, computational biology, and cellular neuroscience. My scientific journey began in 2008 at the Center for Genomic Regulation (Barcelona, Spain), where I applied computational biology to investigate molecular epistasis. I later earned a Ph.D. in Genomics and Bioinformatics from the University of Southampton (United Kingdom), focusing on epigenetic and transcriptomic mechanisms underlying neuropsychiatric disorders. In 2016, I undertook postdoctoral training in Molecular Psychiatry and Genomics at the Icahn School of Medicine at Mount Sinai. Building on this foundation, I joined the Faculty at Mount Sinai in 2019, where I established the A-to-I Editing and Rare Disorders Lab within the Departments of Psychiatry and Genetics and Genomic Sciences. Our lab is a proud member of the Seaver Autism Center for Research and Treatment, the MINDICH Child Health and Development Institute, and the Friedman Brain Institute. Together, we are dedicated to advancing the understanding of brain function and disease through innovative and collaborative research.




Contact: olga.dmitrichenko [@] mssm.edu
As a health scientist and epidemiologist, Olga’s work focuses on suboptimal pregnancy and neurodevelopmental aspects during childhood, linked with neuroimaging. She has worked on the Generation R Cohort at the Erasmus Medical Center in Rotterdam, the Netherlands, and conducted research on eating behaviours in association with brain morphology in children. After joining Icahn School of Medicine at Mount Sinai, her work focuses on maternal immune activation and brain outcomes, where she is also part of the Women’s Mental Health Program. Outside research, she enjoys long walks with friends and art expositions.

- Xuanjia (Sinja) Fan was a Associate Researcher/Lab Manager and is now attending Medical School at Penn State.
- Enrico Mossotto was a Postdoctoral Fellow and is now leading genomic research in Industry
-
Joshua Senior was a research volunteer from Brooklyn Technical High School and is now attending University.
-
Kendall Moore was a SURP student and is currently finishing studies at Santa Clara University studying Neuroscience and Ethnic Studies.
-
Sarah Zipkowitz was a Research Volunteer and is currently attending Binghamton University where she is studying Biology and Judaic Studies on the Pre-Med track.
- Gauri Ganesh was a MSc student and is now a bioinformatician in industry.
- Ariela Buxbaum Grice was an Associate Researcher and is now a PhD graduate student.
Our Research
Genomics of brain development and neurodevelopmental disorders
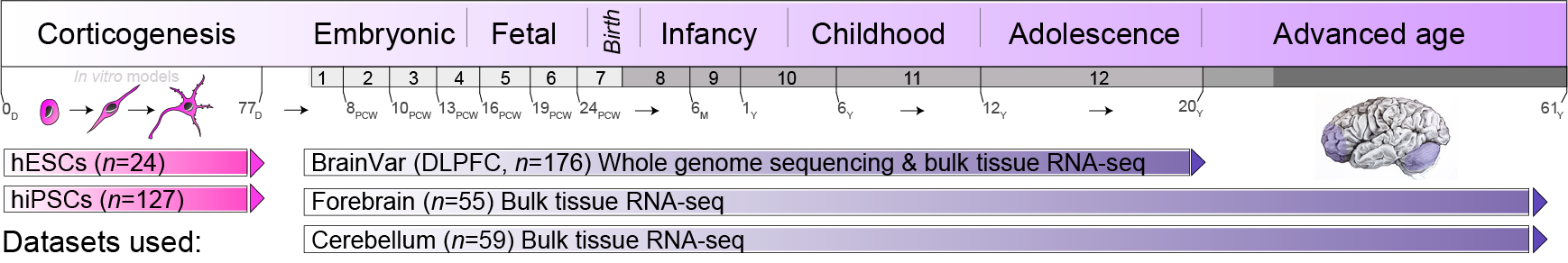
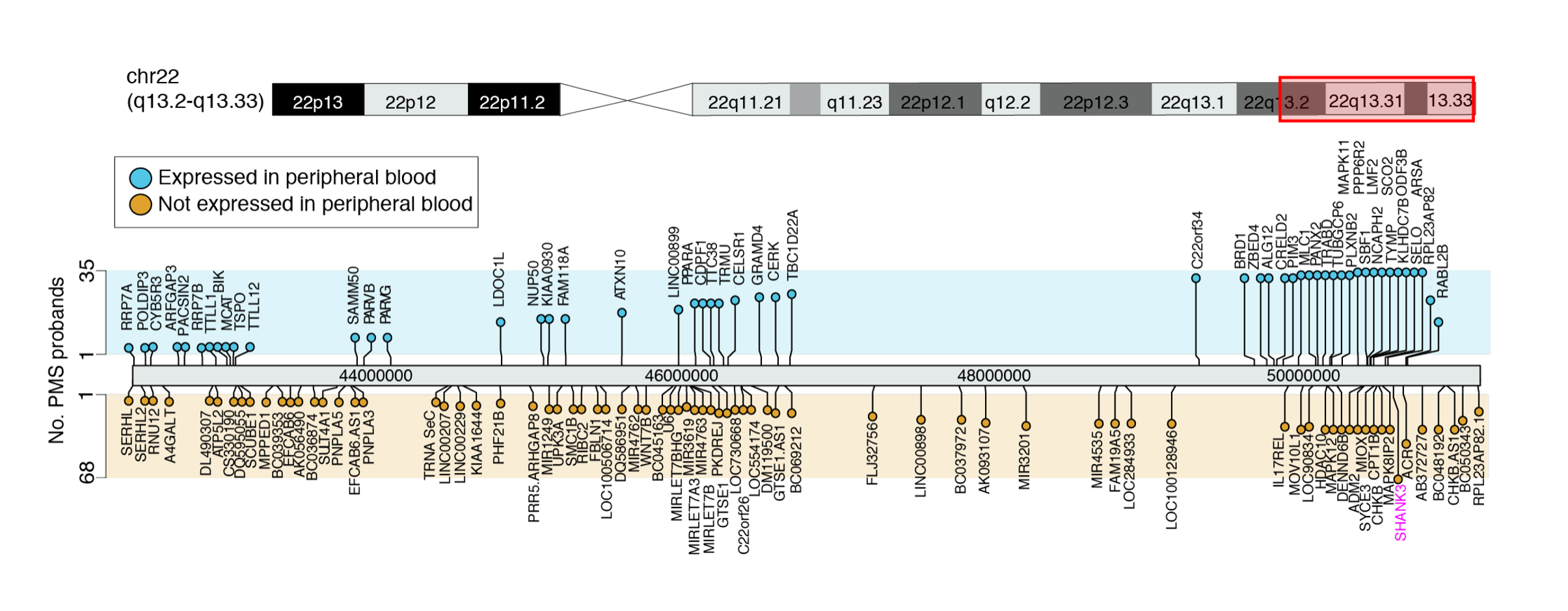
Our work pioneers large-scale genomic investigations of human cortical developmental, combing genetics, transcriptomics and proteomics (see here and here). These studies chart the complex spatiotemporal and genetic regulation of gene expression and protein abundance, their functions and timely interactions across fetal and postnatal development. We have also led studies dissecting post-transcriptional A-to-I modifications and their roles in supporting the functional diversity of human brain development (see here). In addition, our lab is part of the Autism Sequencing Consortiums (ASC), dedicated to dissecting the genetic architecture of ASD through large-scale whole exome sequencing and de novo variant discovery (see here). We are also part of the Seaver Autism Center for Research and Treatment at Mount Sinai, where we led a number of studies focused on rare monogenic subtypes of autism (see here, a report of transcriptional alterations in hiPSC-derived neurons as a consequence of 22q13.3 sequence variants and deletions in participants with Phelan-McDermid syndrome, and here, a report on ketamine-induced transcriptional responses in children with ADNP syndrome).
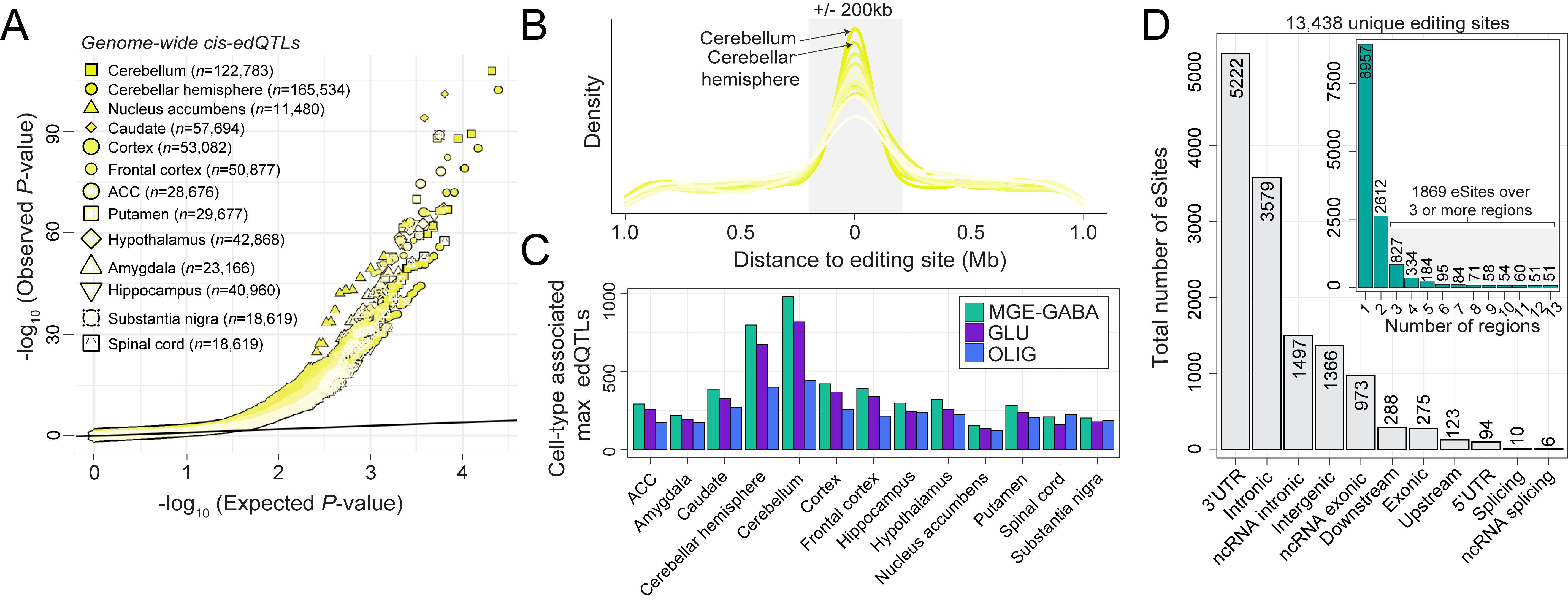
Mapping the regulation and function of A-to-I editing in brain health and disease
Another component of our work is focused on elucidating highly regulated adenosine-to-inosine (A-to-I) editing sites in the human brain both with the main goal to prioritize RNA editing sites for functional, mechanistic, and RNA therapeutic investigation. Currently, we are tackling aspects of RNA editing regulation across brain development (see here), major brain cell types and their genetic regulation (see here) and dysregulation in neurological disorders (see our work in schizophrenia here). We have also investigated A-to-I editing biology in living human brain tissue (see here). Currently, we are focused on three central areas:
- First, we aim to uncover A-to-I sites implicated in neurodevelopmental and neurodegenerative disorders using large-scale RNA-seq and snRNA-seq from postmortem brain samples and neuronal cell systems.
- Second, we seek to examine the functional impact of these RNA editing sites on protein function and cellular phenotypes, and we are specifically focused on sites encoding glutamatergic receptors, ion channels and pumps.
- Third, we aim to develop site-directed RNA editing approaches for therapeutic correction of disease-related RNA editing sites as well as therapeutic repair of highly penetrant, rare genomic point mutations at the mRNA level.
Immuno-genetics of neurodevelopmental disorders: biomarkers and treatments
We have investigated the immunobiology of major neurological disorders for the past 10 years (see a few examples here, here, here, here). In the Seaver Autism Center, we are leveraging this expertise to dissect the immunogenetics and immunobiology of children with rare developmental disorders. Our research in the Center surveys peripheral immune responsivity and function in children with rare syndromic subtypes of autism using high-throughput genomic and cellular techniques, including scRNA-seq, metabolomics, proteomics and cytometry by time of flight. This work has uncovered novel immunodeficiencies that underlie recurrent infections in rare disorders that are typically studied in the context of the central nervous system, thereby providing avenues for biomarker development and new therapeutic interventions to tackle common immune-related phenotypes. Our work has also described novel molecularly defined genetic subtypes in both ADNP syndrome (see here) and Phelan McDermid Syndrome (see here).
Algorithm Development
We are also interested in solving emerging biological and algorithmic problems, which arise from our studies and others, such as: a) comparative transcriptomics and proteomics; b) modelling transgenerational effects across generations; c) predicting cellular frequencies from heterogeneous biological tissue; d) multi-modal integrative deep machine-learning applications; e) modelling RNA-editing in from heterogeneous RNA-sequencing data; f) gene network reconstruction and multi-modal omic data integrations. In addition to generating new data in support of these aims, we also use just about any high-throughput data we get our hands on in the public domain, which can ultimately be translated into better understanding biology.
Check some of our Rshiny apps:
The Seaver Autism Center for Research and Treatment

The Breen Lab is part of the Seaver Autism Center for Research and Treatment at the Icahn School of Medicine at Mount Sinai. The Seaver Autism Center is a fully integrated, translational research center that leads progressive studies and provides personalized care to individuals with autism and related rare disorders. As a team, we are dedicated to discovering the biological causes of autism and developing breakthrough treatments.
As a part of the Seaver Center, our lab is primarily focused on understanding the cellular and molecular consequences underlying rare syndromic forms of autism, including Phelan McDermid-Syndrome (SHANK3), ADNP syndrome, and FOXP1 syndrome. We also collaborate with groups in the Center that generate genetically modified animal models for these genes as well as human iPSC-derived neurons, which we profile using functional genomic tools. Learn more about the Seaver Center here: https://icahn.mssm.edu/research/seaver
Follow the Seaver Autism Center on Twitter:
Select lab publications
- Divergent landscapes of A-to-I editing in postmortem and living human brain. Nature Communications. (2024).
- Spatiotemporal and genetic regulation of A-to-I editing throughout human brain development. Cell Reports. (2022).
- Cellular and genetic drivers of RNA editing variability in the human brain. Nature Communications. (2022)
- Modeling gene × environment interactions in PTSD using human neurons reveals diagnosis-specific glucocorticoid-induced gene expression. Nature Neuroscience. (2022)
-
Episignatures Stratifying Helsmoortel-Van Der Aa Syndrome Show Modest Correlation with Phenotype. The American Journal of Human Genetics. (2020).
- Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. (2020)
- Global landscape and genetic regulation of RNA editing in cortical samples from individuals with schizophrenia. Nature Neuroscience. (2019).
- Differential transcriptional response to glucocorticoid activation in cultured blood immune cells: a novel approach to PTSD biomarker development. Translational Psychiatry. (2019) .
- Large-scale proteomic analysis of human prefrontal cortex identifies proteins associated with cognitive trajectory in advanced age.Nature Communications (2019).
- Temporal proteomic profiling of postnatal human cortical development. Translational Psychiatry (2018).
- Gene expression in cord blood links genetic risk for neurodevelopmental disorders with prenatal maternal distress and adverse childhood outcomes. Brain, Behavior, and Immunity (2018).
Available Positions
We strive to maintain an open-minded, creative and productive research environment, and we are fully committed to foster the growth and development of trainees in the wet and dry labs. If you are interested in applying for a position, please send the following information to michael.breen@mssm.edu
• Complete curriculum vitae, including a list of publications.
• A very brief summary of current work and research interests.
• Contact information for two references.
Contact Us
Contact: Depts. Psychiatry, Genetics and Genomic Sciences
ICAHN BUILDING 14-26
Icahn School of Medicine at Mount Sinai
1425 Madison Avenue, Box 1498
New York, NY 10029