Our Research
The Bevacqua Lab studies human diabetes, with the goal of expanding and improving human pancreatic beta cell mass function for the 400+ million people in the world with diabetes. More specifically, we focus on pancreatic islet biology, attempting to understand the regulatory mechanisms driving postnatal human islet maturation and the genetic basis for their dysregulation in disease, findings that will be applied towards β-cell regeneration and replacement therapies. Questions of the Bevacqua lab include: “What transcription factors are required for proper maturation of human islet cells?” “What is the role of non-coding regulatory elements in modulating mature function of islets?” “What mechanisms are perturbed by risk variants linked to diabetes?” And, “What are the roles of chromatin modification and external signals in regulating this terminal maturation?”
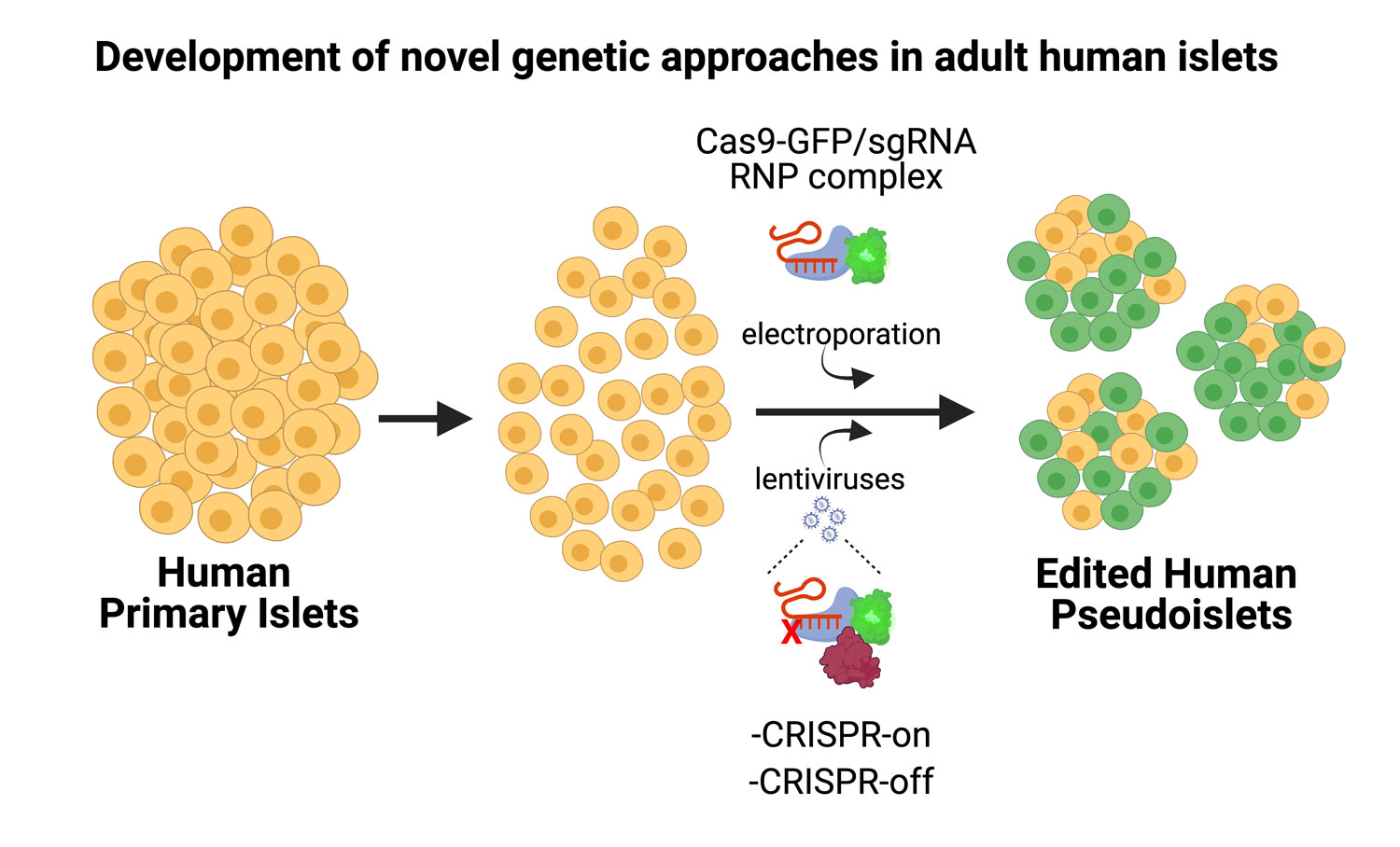
Our recent development of genetic systems in primary human islet organoids, termed “pseudoislets”, now provides unprecedented methods to investigate mechanisms regulating function of mature islets. Our approaches will include implementation of novel genomic approaches in vivo to reverse diabetes, and will open new possibilities for the treatment of patients with diabetes.
General Overview of Research
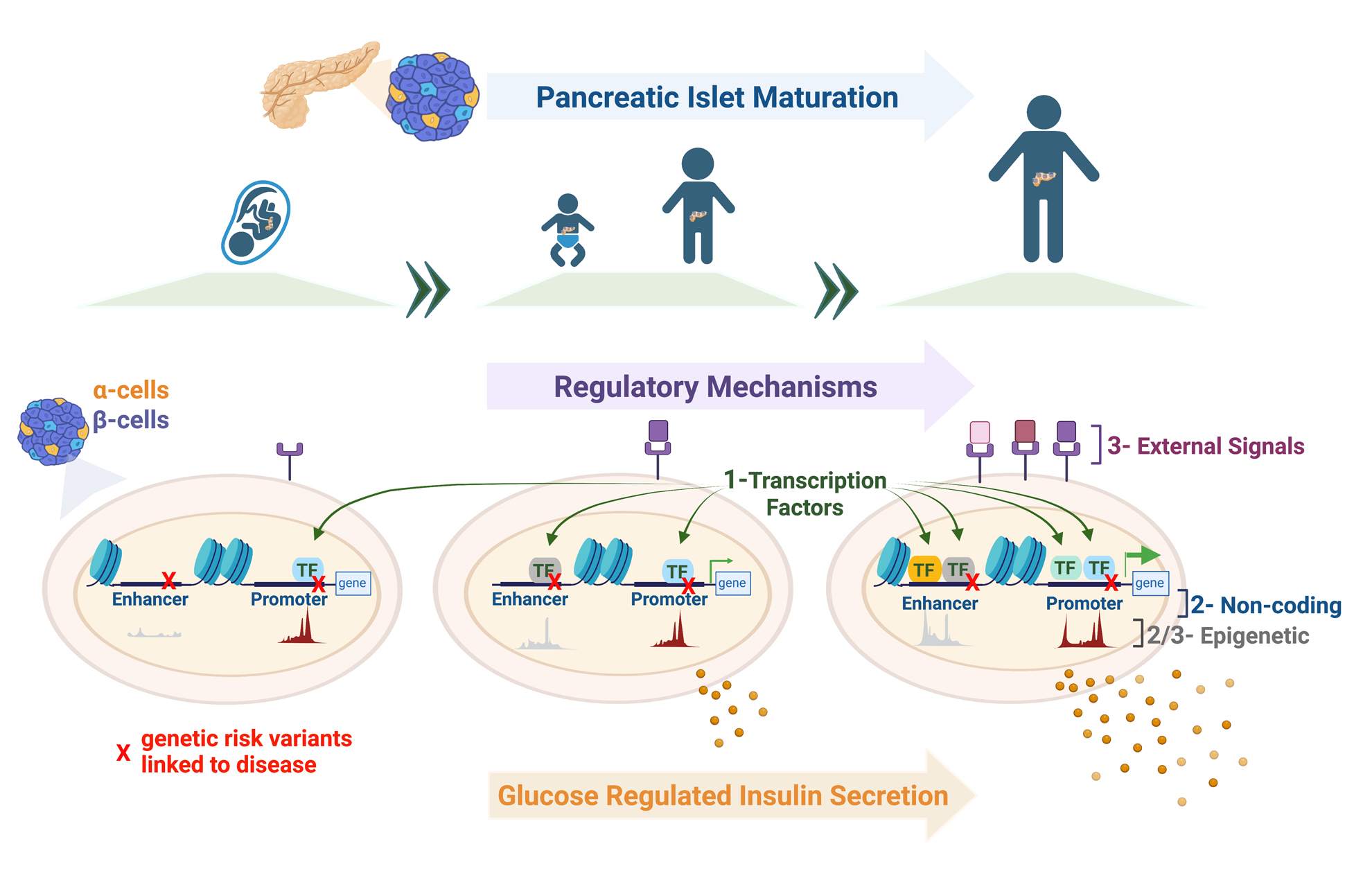
Research Methods: Innovative Systems to Study Human Islet Biology
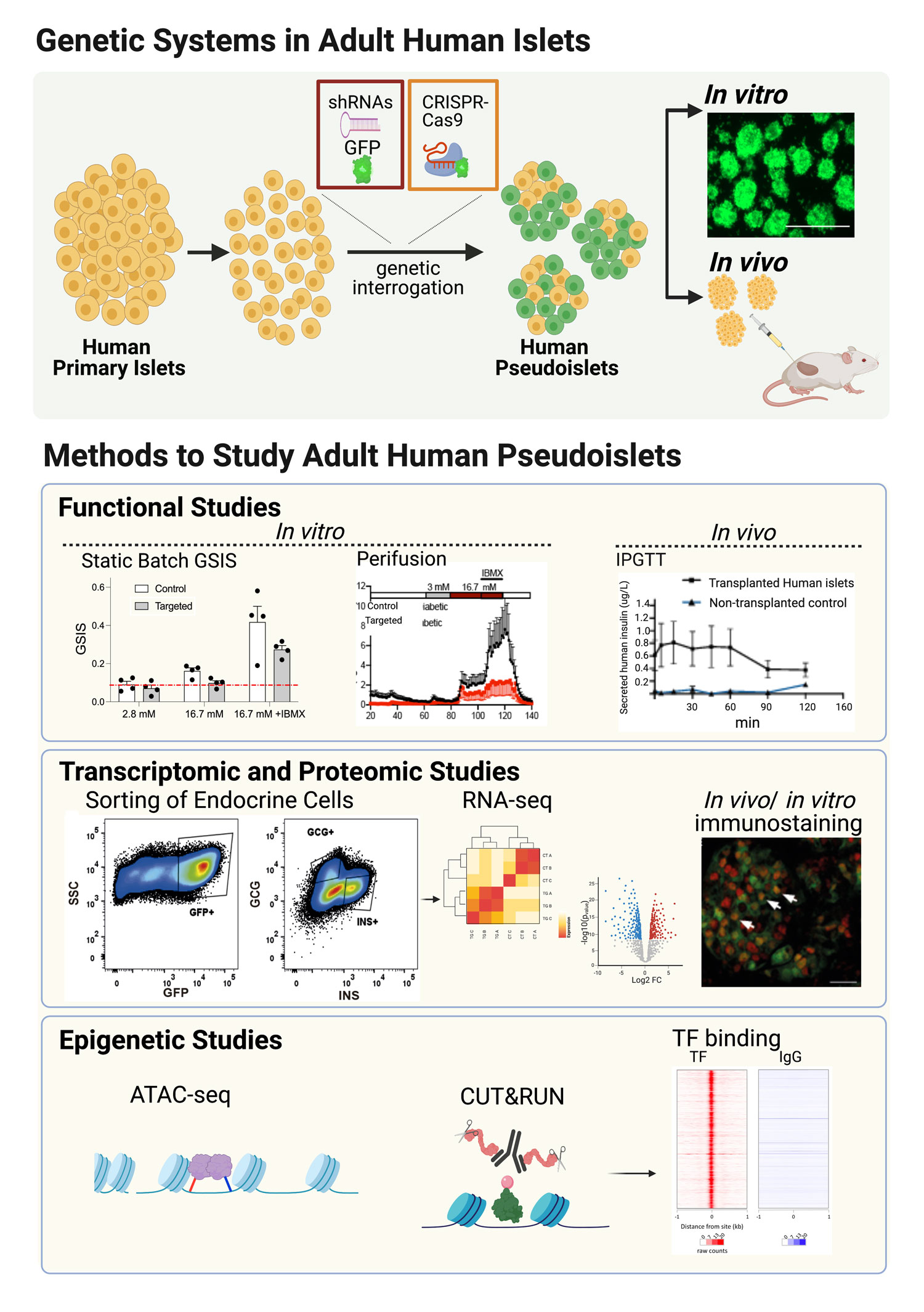
Projects
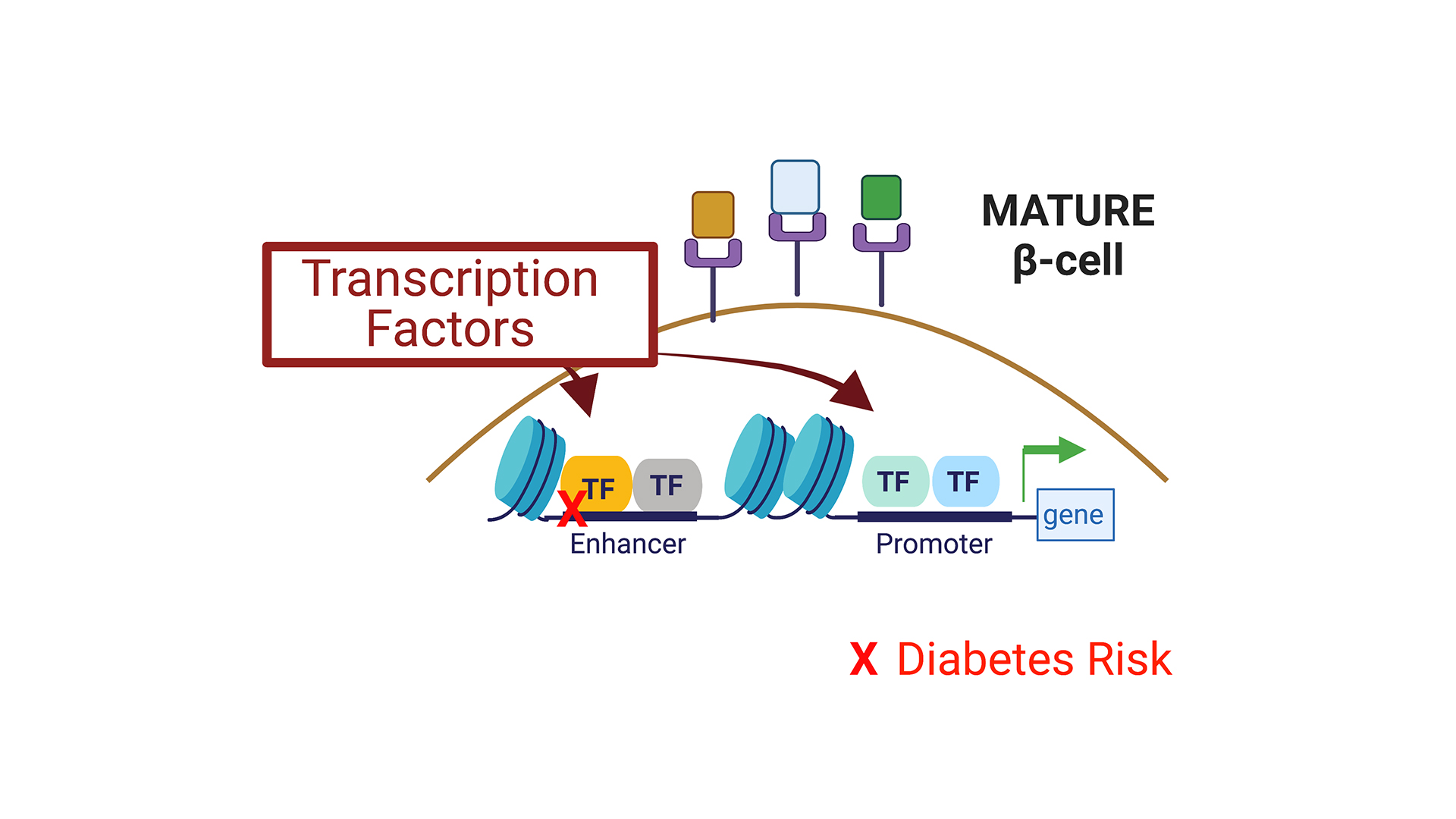
Project 1. Identification of transcription factors and pathways driving pancreatic islet maturation.
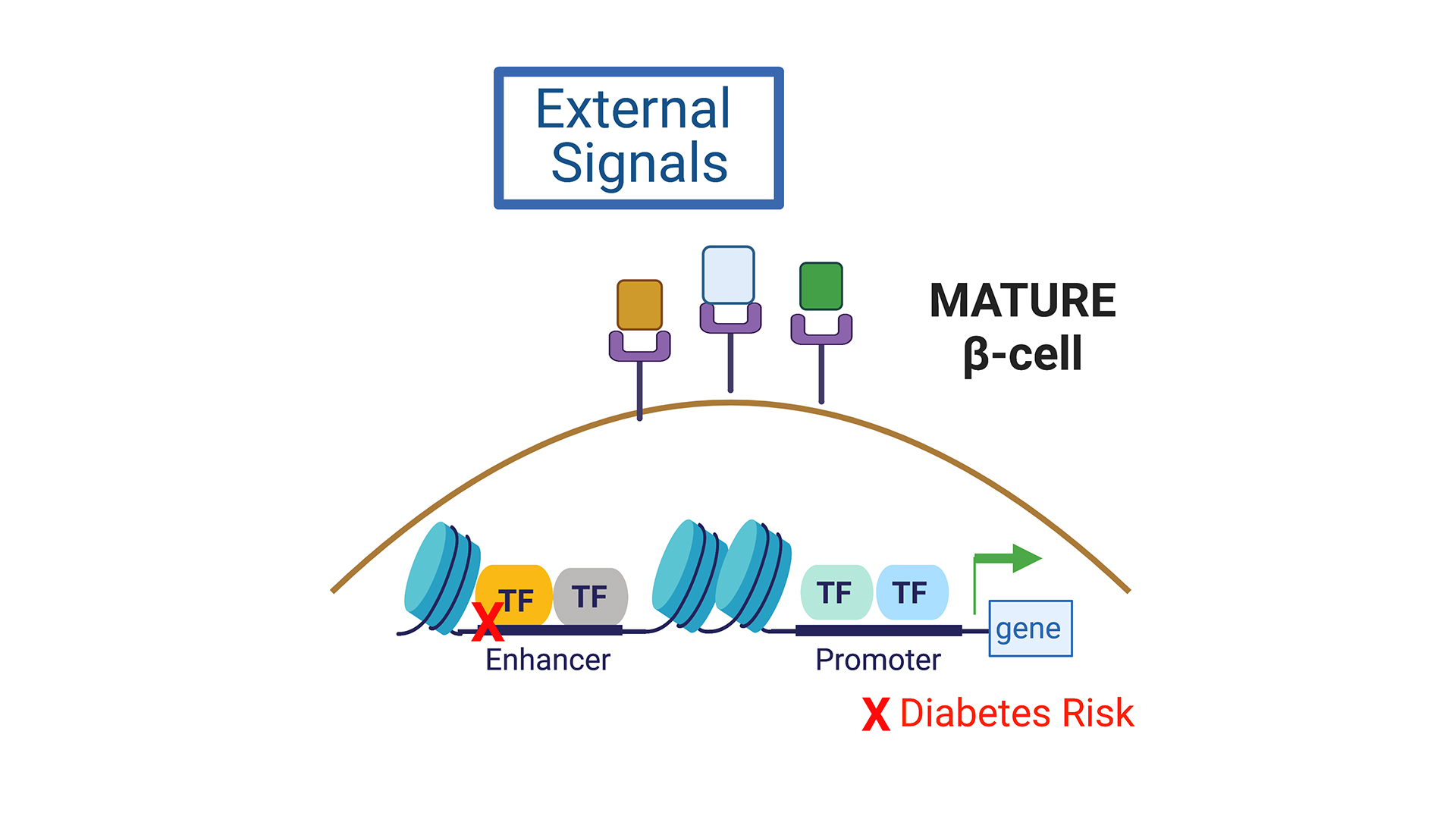
Transcription factors (TFs) play a critical role in regulating β-cell maturation, as evidenced by studies of human monogenic diabetes mellitus, and impaired β-cell function resulting from mutations in multiple TF genes. In recent years, many human-specific TFs differentially expressed across development and disease states were identified, reflecting efforts to reliably procure human islets for research, and advances in NGS-based technologies for small cell number samples. Using genetic systems in primary human pseudoislets, the Bevacqua lab studies mechanisms regulated by TFs dynamically expressed in human islet b- and a-cells. We hypothesize that TFs showing dynamic expression during a critical window of developmental maturation, and linked to diabetes disease risk, could drive the maturation of these cells. Our studies should elucidate new genetic and chromatin regulatory networks that govern human islet cell function, and provide a much-sought ‘roadmap’ for guiding current world-wide islet cell replacement and regeneration efforts.
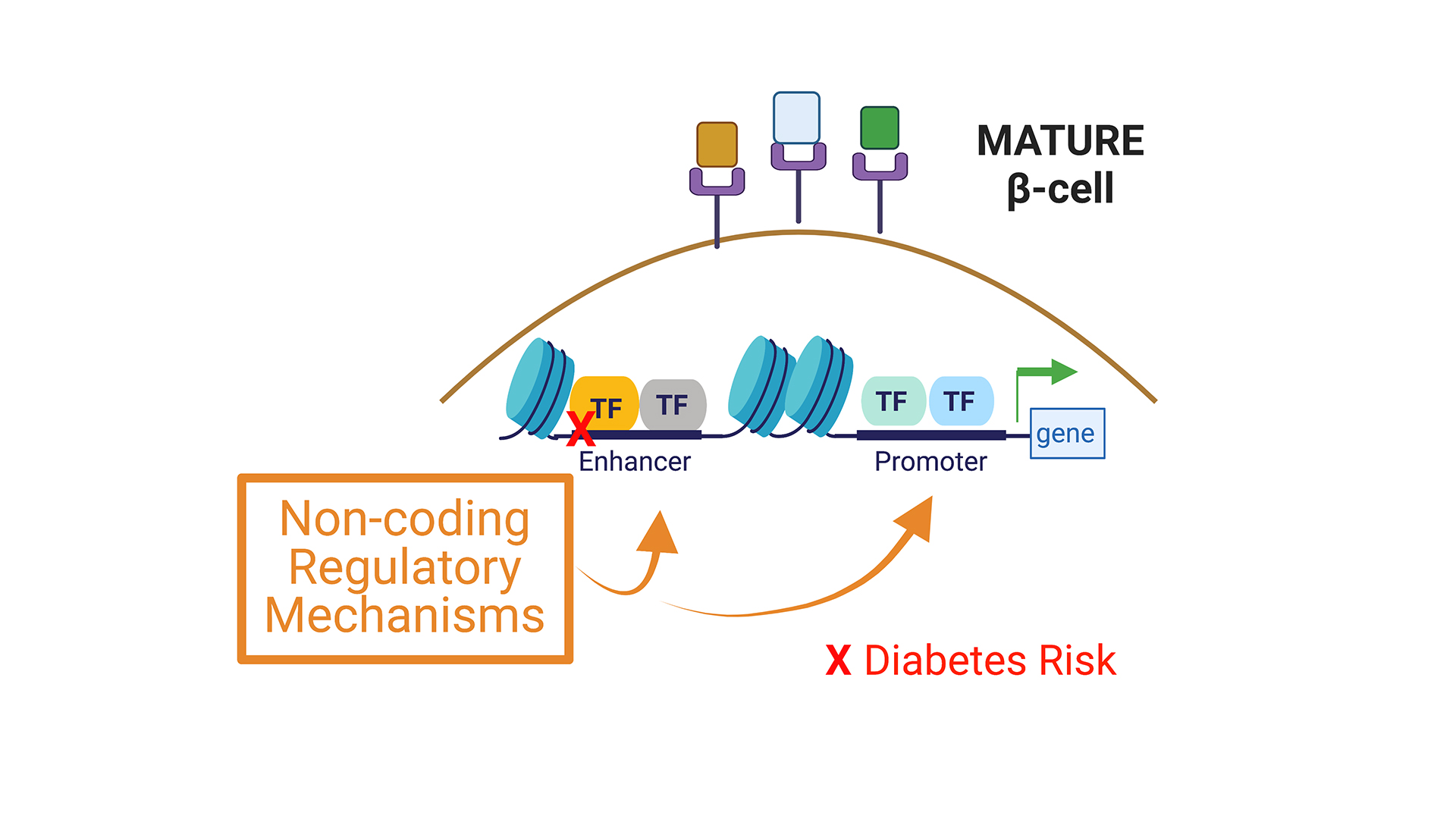
Project 2. Deciphering non-coding regulatory mechanisms driving islet cell maturation and dysregulation.
The vast preponderance of genetic risk for diabetes mellitus (at >400 risk loci) maps to non-coding DNA, mostly in presumptive regulatory regions. In other cell systems such as neurons, licensing and decommissioning of thousands of cis-regulatory enhancer elements mediates postnatal maturation. Whether in neurons or islets, we still lack understanding of the target genes of most of these enhancers, and of their dynamic age-dependent regulation. Recent developments by Dr. Bevacqua, including CRISPR-based systems to study non-coding regulatory sequences in primary non-dividing cells (Bevacqua et al. 2021a) and CUT&RUN -Cleavage Under Targets and Release Using Nucleases- from low cell inputs, such as those available from human islets (Bevacqua et al. 2021b) introduce unprecedented possibilities to identify and study the function of these regulatory elements in a bona fide islet context. Identification of enhancer regions that are differentially accessible throughout islet maturation and disease will contribute to our understanding of the genetic and epigenetic mechanisms that contribute to islet β-cell dysfunction in diabetes.
Project 3. Elucidating external signals regulating pancreatic islet maturation.
Postnatal maturation of human islet cells occurs over a prolonged period from birth to early adulthood, developmental stages that coincide with changes in diet, nutrient metabolism, and hormone levels. However, the signaling basis of this fascinating example of cellular development remains poorly understood. Based on recent publications identifying differential regulation of feeding-induced enhancers with age in human islet cells, we hypothesize that nutrient- and hormone-linked islet enhancers might be enriched for motifs recognized by TFs that show differential expression with age, including those “priming” functional islet maturation. We plan to study these mechanisms using reverse genetics in primary human islets and mouse models with mutations in signal-linked-TF binding motifs, combined with exposure to environmental stimuli. These studies could provide valuable information about how to generate functionally mature β-cells and to restore function to diseased β-cells.
Project 4. Development of novel in vitro and in vivo genomic approaches to reverse diabetes.
Technological advances foster discovery. We are currently developing novel CRISPR approaches, including using electroporation of ribonucleoproteins (RNPs) to delete non-coding regulatory DNA elements, without integration of lenti-vectors. This approach has multiple advantages, including simplicity, scalability, avoidance of lentiviral integration or toxicity, and adaptation for multiplexing, relevant to diabetes modeling. In addition, we are working to adapt additional dCas9- based approaches, that will allow the study of epigenetic mechanisms relevant to maturation and disease.
Current Members
Dr Romina Bevacqua, Assistant Professor
Department of Medicine, Division of Endocrinology, Diabetes and Bone Disease
Member of the Diabetes Obesity Metabolism Institute and the Regenerative Biology and Stem Cell Institute
Dr. Romina Bevacqua is an Assistant Professor in the Department of Medicine, Division of Endocrinology, Diabetes and Bone Disease, and member of the Diabetes, Obesity and Metabolism Institute (DOMI) and the Regenerative Biology and Stem Cell Institute, at Mount Sinai.
Dr. Bevacqua received her PhD in Buenos Aires University, Argentina. As a postdoctoral fellow in the laboratory of Professor Seung Kim, at Stanford University, her research focused on human islet biology.
Her research career vision is to perform transformative research on human diabetes, with the specific goal of expanding and improving human pancreatic beta cell mass function for the 400+ million people in the world with diabetes.
Contact: romina.bevacqua@mssm.edu
Dr Emilio Merheb, Senior Associate Researcher
Department of Medicine, Division of Endocrinology, Diabetes and Bone Disease
Member of the Diabetes Obesity Metabolism Institute and the Regenerative Biology and Stem Cell Institute
Emilio Merheb was born and raised in Byblos, Lebanon. At the age of eighteen, he was offered a scholarship to pursue his undergraduate degree at the Lebanese American University. He completed his PhD and postdoctoral training at Albert Einstein College of Medicine, where his work focused on investigating the mechanism of a rare neurodegenerative disease, Pol III-related Leukodystrophy. Notably, his work was published in high impact peer reviewed journals, including PNAS, Cell Metabolism and Cells. Emilio’s dedication to science paid off with multiple awards including the Julius Marmur Award and the Graduate Student Council Service Award. In his free time, he enjoys sports, music and spending time with his family and friends.
Contact: emilio.merheb@mssm.edu
Dr Emilio Merheb, Senior Associate Researcher
Department of Medicine, Division of Endocrinology, Diabetes and Bone Disease
Member of the Diabetes Obesity Metabolism Institute and the Regenerative Biology and Stem Cell Institute
Emilio Merheb was born and raised in Byblos, Lebanon. At the age of eighteen, he was offered a scholarship to pursue his undergraduate degree at the Lebanese American University. He completed his PhD and postdoctoral training at Albert Einstein College of Medicine, where his work focused on investigating the mechanism of a rare neurodegenerative disease, Pol III-related Leukodystrophy. Notably, his work was published in high impact peer reviewed journals, including PNAS, Cell Metabolism and Cells. Emilio’s dedication to science paid off with multiple awards including the Julius Marmur Award and the Graduate Student Council Service Award. In his free time, he enjoys sports, music and spending time with his family and friends.
Contact: emilio.merheb@mssm.edu
Selected Publications
Rottner AK, Ye Y, Navarro-Guerrero D, Rajesh V, Pollner A, Bevacqua RJ, Yang J, et al. A genome-wide CRISPR screen identifies CALCOCO2 as a regulator of beta cell function influencing type 2 diabetes risk. Nature Genetics 2022; https://doi-org/10.1038/s41588-022-01261-2
https://pubmed.ncbi.nlm.nih.gov/36543916/
Bevacqua RJ, Dai X, Lam JY, Gu X, Friedlander MSH, Tellez K, Miguel-Escalada I, Dominguez AA, Qi LS, Ferrer J, MacDonald PE, Kim SK. CRISPR-based gene editing in primary human pancreatic islet cells. Nature Communications 2021;12(1):2397.
Perspective: A commentary by Romina Bevacqua. Behind the Paper: CRISPR-based genome editing in primary human pancreatic islet cells. Nature Portfolio Bioengineering Community. 2021. Weblink: https://bioengineeringcommunity.nature.com/posts/crispr-based-genome-editing-in-primary-human-pancreatic-islet-cells
Highlight: Latest HIRN Science. HIRN Newsletter. April 2021. Weblink: https://myemail.constantcontact.com/HIRN-April-2021-Newsletter.html?soid=1125971089264&aid=vYWEZRRUDTA
https://pubmed.ncbi.nlm.nih.gov/33893274/
Bevacqua RJ, Lam JY, Peiris H, Whitener RL, Kim S, Gu X, and Kim SK. SIX2 and SIX3 coordinately regulate functional maturity and fate of human pancreatic b-cells. Genes and Development 2021;35(3-4):234-249.
Journal Highlight: Genes and Development Cover, February 1, 2021:35(3-4)
https://pubmed.ncbi.nlm.nih.gov/33446570/
Wu CT, Hilgendorf KI, Bevacqua RJ, Hang Y, Demeter J, Kim SK, Jackson PK. Discovery of ciliary G protein-coupled receptors regulating pancreatic islet insulin and glucagon secretion. Genes and Development 2021;35(17-18):1243-1255.
https://pubmed.ncbi.nlm.nih.gov/34385262/
Wu CT, Lidsky PV, Xiao Y, Lee IT, Cheng R, Nakayama T, Jiang S, Demeter J, Bevacqua RJ, Chang CA, Whitener RL, Stalder AK, Zhu B, Chen H, Goltsev Y, Tzankov A, Nayak JV, Nolan GP, Matter MS, Andino R, Jackson PK. SARS-CoV-2 infects human pancreatic β cells and elicits β cell impairment. Cell Metabolism 2021;33(8):1565-1576.e5.
https://pubmed.ncbi.nlm.nih.gov/34081912/
Friedlander MSH, Nguyen V, Kim S*, Bevacqua RJ*. Pancreatic pseudoislets: an organoid archetype for metabolism research. Diabetes 2021;70(5):1051-1060.
https://pubmed.ncbi.nlm.nih.gov/33947722/
Kim S, Whitener RL, Peiris H, Gu X, Chang CA, Lam JY, Camunas-Soler J, Park I, Bevacqua RJ, Tellez K, Quake SR, Lakey JRT, Bottino R, Ross PJ, Kim SK. Molecular and genetic regulation of pig pancreatic islet cell development. Development 2020;147(6):dev186213.
Journal Highlight: Pancreatic development in the pig. Development 2020;147(6):e0604.
https://pubmed.ncbi.nlm.nih.gov/32108026/
Funding and Awards
2023 NIDDK-HIRN CHIB New Investigator Gateway Awards for Collaborative T1D Research (R03)
2023 Biomedical Laureate
2018-2021 JDRF Postdoctoral Scholar

Positions
The Bevacqua Lab is always interested in hearing from curious, motivated potential students, post-doctoral trainees and research associates who are excited in transformative research on human diabetes and pancreatic islet biology.
Formal announcements of positions will be posted here in due course but informal enquires are welcome anytime. Please send your CV and a covering letter outlining your motivation for applying to romina.bevacqua@mssn.edu.
Current Open Positions
We are currently looking for a Postdoc and Grad Student to join the lab. Email Dr. Bevacqua if interested!
The Bevacqua Lab





