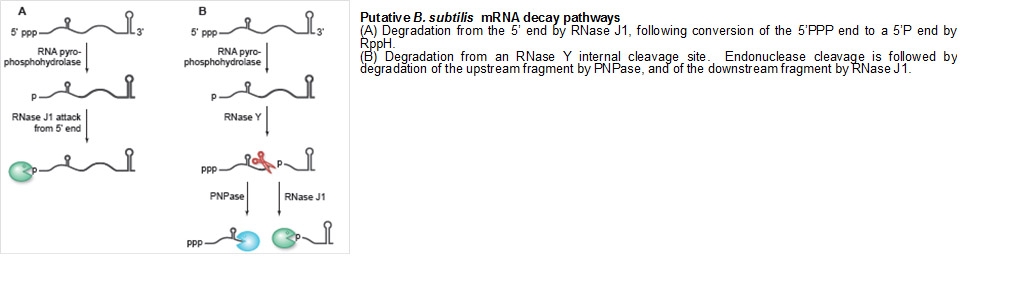
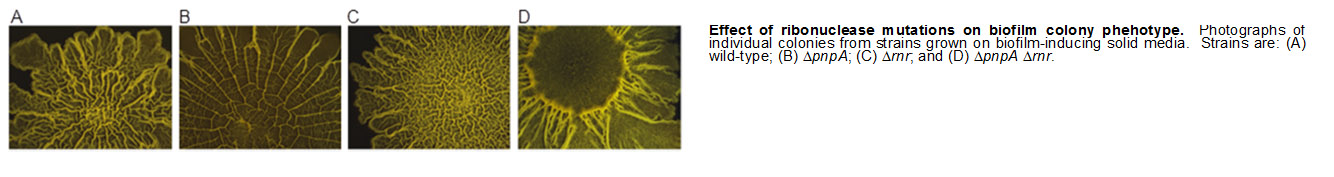
Decay of bacterial mRNA is an essential function, needed constitutively to regenerate ribonucleotide pools and in a regulated fashion to confer flexibility on gene expression programs. Relative to our understanding of the processes of transcription and translation, much less is known about how mRNA turnover is accomplished, and much of what we do know comes from studies in Escherichia coli, the Gram-negative model organism. We have been studying mRNA decay and processing in the Gram-positive bacterium, Bacillus subtilis, which is so far the best understood organism other than E. coli in terms of the ribonucleases involved in mRNA decay. Over the years, our lab has discovered many genes encoding ribonuclease activities, and has characterized the bacterial mRNA decay process, using a handful of “model” mRNAs as representatives of the transcriptome. We now routinely use NextGen sequencing technology to assay all expressed mRNAs at once, employing newly-constructed mutant strains to determine the role that particular activities play in mRNA turnover. In addition to studying individual proteins, we are beginning to investigate a protein complex called the “degradosome,” which comprises several proteins that may function jointly in achieving efficient mRNA turnover.