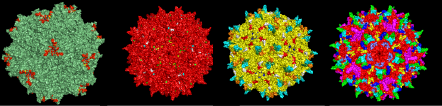
Together, the various serotypes and variants of AAV that have been described can transduce a wide array of cells. Nevertheless, certain cells are refractory to transduction. In addition, for many gene therapy applications, it is desirable to transduce only specific cell types. Over the past few years, several methods—biochemical as well as genetic—have been developed to expand and/or restrict AAV tropism.
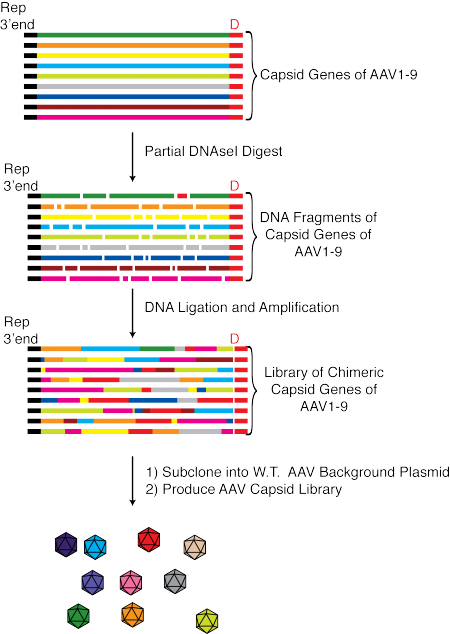
Among the most popular approaches to select AAV variants with improved properties is directed evolution using libraries of AAV variants with diverse AAV capsids. High complexity AAV libraries can be created by a number of methods, e.g. by inserting degenerate peptides into hypervariable surface loops, via the use of error-prone PCR to introduce point mutations into the AAV capsid or through so-called capsid shuffling (see figure below).
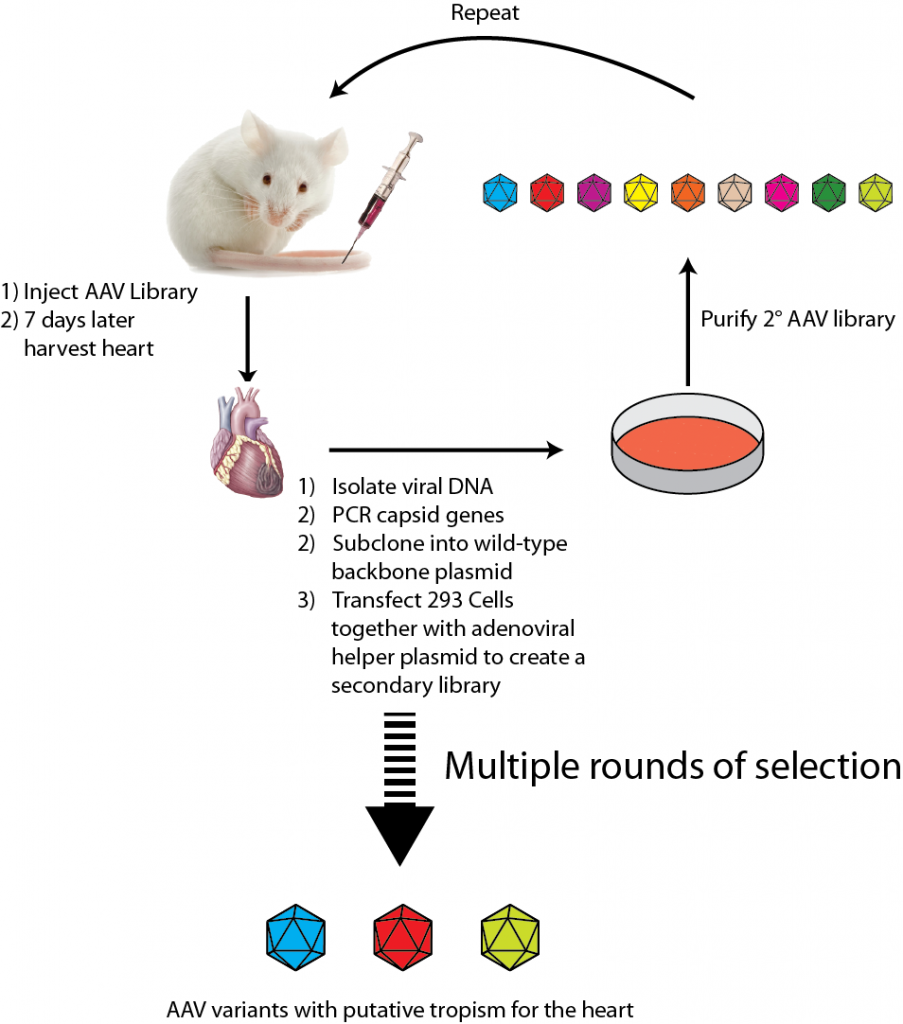
Directed evolution is a particularly widely used approach that has been used to select AAV variants with altered tropism. An experimental outline of using directed evolution to select AAV variants that have tropism for the heart is shown below.
Challenges in AAV Capsid Library Production
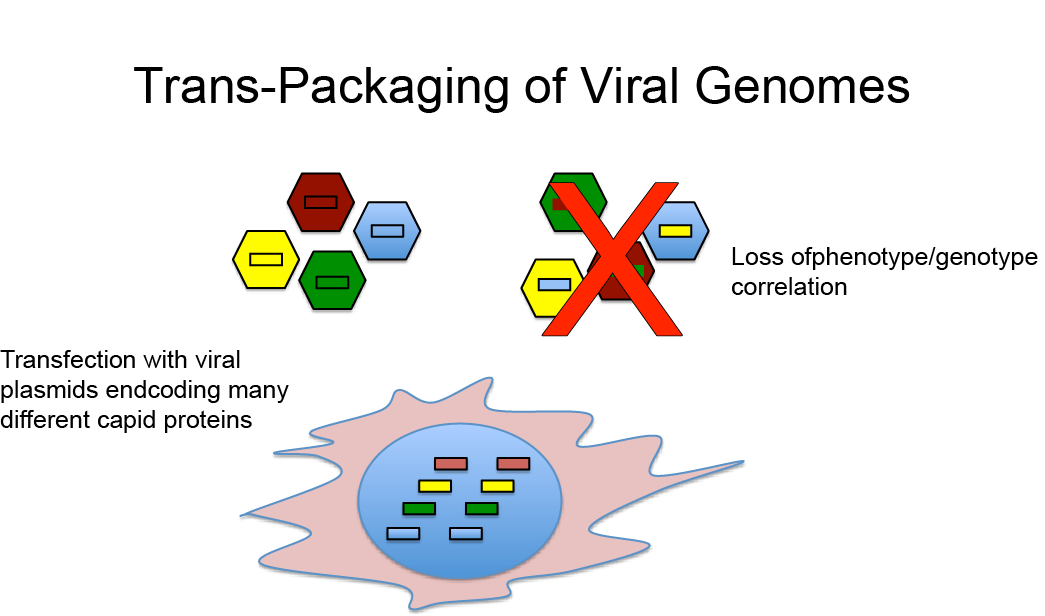
Potential Loss of Genotype/Phenotype Correlation During AAV Library Creation (Nonnenmacher et al., 2015)
AAV libraries with diverse capsids are in general produced by the co-transfection of HEK293 cells with a library of plasmids encoding the Rep proteins of AAV2, which are necessary for DNA replication and DNA packaging, and a variety of AAV capsid variants flanked by two inverted terminal repeats of AAV2 together with a helper plasmid that encodes all the necessary adenoviral helper functions (except E1A, which is provided by the HEK293 cells).
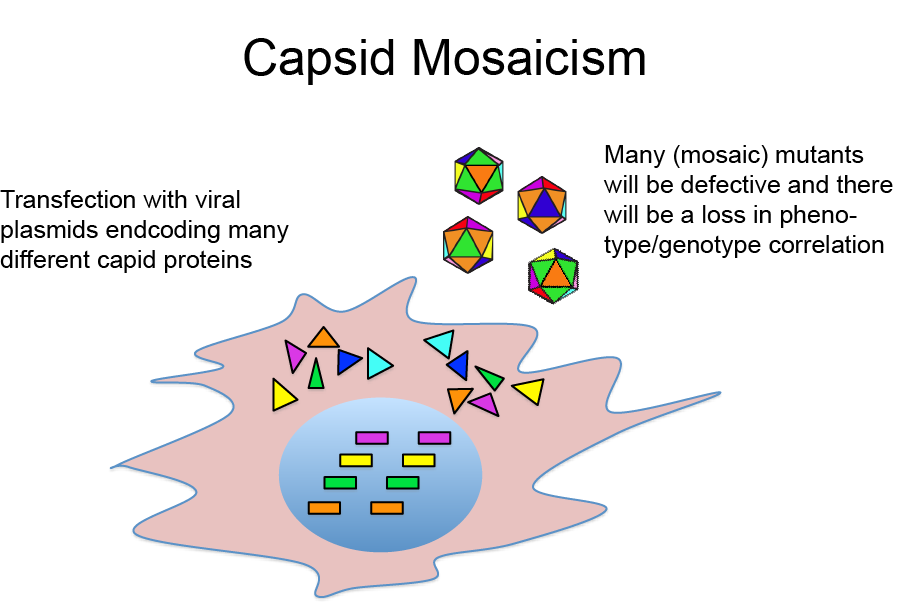
A potential complication with this approach is that because plasmids encoding many different capsid sequences are introduced into a single cell, potentially many different viral genomes and capsid proteins will be produced in the same cell. This can be problematic for two reasons:
- The simultaneous expression of many different capsid proteins in the same cell can lead to viral mosaics, i.e. virions composed of more than one type of capsid proteins. It is likely that many of those mosaic virions are non-infective. Moreover, because each virion can only package one viral genome encoding one specific capsid protein, this results in a loss of capsid phenotype viral genotype correlation.
- Even if the virions produced in a cell with multiple AAV plasmids are composed of only one capsid protein, it is still possible that a genome encoding capsid variant A is encapsidated into a viral capsid composed of capsid proteins B again resulting in a loss of phenotype genotype correlation.
Importantly, the loss of a strict phenotype/genotype correlation is disastrous for the use of directed evolution to select AAV variants with improved properties because the successful use of directed evolution is absolutely dependent on the fact that the capsid proteins of a virions are encoded by the encapsidated viral genome.
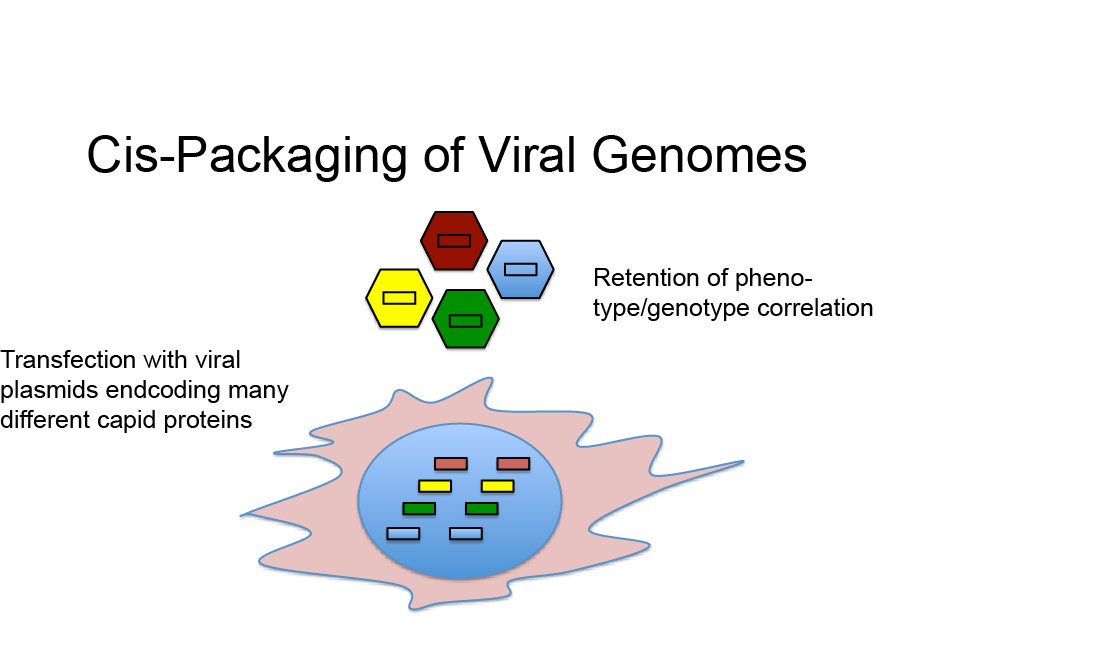
High Phenotype Genotype Correlation in AAV Libraries
In view of the above considerations it appears surprising that directed evolution with AAV libraries could be used successfully to isolate AAV variants with novel properties, e.g. alternate tropism or increased resistance to pre-existing neutralizing antibodies. However, although being counterintuitive, we were recently able to demonstrate that the vast majority of AAV capsids in AAV libraries likely harbor a genome encoding a matching capsid protein sequence, which offers a potential answer to this apparent paradox (Nonnenmacher et al., 2015).
Current efforts are underway to understand this puzzling phenomenon. In addition, our lab is actively studying approaches to create improved AAV libraries that enhance the chances of being able to isolate AAV variants with the desired properties.
Reference