Identifying tumor-intrinsic signaling pathways that restrict anti-tumor immunity in hepatocellular carcinoma
Nivolumab, a novel PD-1 immune checkpoint inhibitor, has shown unprecedented results in a phase II clinical trial in hepatocellular carcinoma, a tumor type that is refractory to other targeted therapies, and has been granted accelerated FDA approval. Unfortunately, the determinants of sensitivity and resistance to this immunotherapy have not been established yet. Here, by combining the use a novel mouse model of HCC immune surveillance, human HCC samples, and transcriptional and immune profiling, we will establish the tumor intrinsic signaling pathways that promote immune escape in HCC, the underlying mechanisms of immune escape, and their effects on response to anti-PD-1 therapy, providing the rationale for improved patient selection and strategies to overcome resistance.
Overcoming resistance to anti-PD-1 immunotherapy in hepatocellular carcinoma
Hepatocellular carcinoma (HCC) represents a major health problem, causing more than 700,000 deaths annually worldwide. Nivolumab, a novel immune checkpoint inhibitor, is currently the most promising therapeutic strategy for HCC patients; however, most patients do not respond, indicating the existence of mechanisms that drive resistance and highlighting the urgent need to identify biomarkers for optimal patient selection and strategies to overcome resistance. Here, by combining a sophisticated mouse model of liver cancer and human HCC samples, we will identify novel mechanisms of intrinsic and acquired resistance to nivolumab. Our project will provide patient selection biomarkers and therapeutic strategies to overcome resistance to nivolumab, and establish a pipeline that can be applied to other immunotherapy drugs or to other tumor types.
Identifying novel combination therapies for liver cancer
Hepatocellular carcinoma (HCC) represents a major health problem, causing more than 700,000 deaths annually worldwide. Although treatment of HCC has greatly improved over the last decades, most HCC patients diagnosed at advanced stages are ineligible for curative ablative therapies such as liver resection or transplantation. The multikinase inhibitor sorafenib was recently approved as the first systemic treatment for patients with advanced HCC, suggesting that targeted therapies could be beneficial in this cancer. However, clinical experiences have revealed that in the most common types of solid tumors, including HCC, progressive lose of efficacy and intrinsic and acquired resistance against molecular therapies are inevitable, indicating that treatment with a single agent is condemned to failure. It is therefore clear that we must move away from a single-target view and toward a paradigm in which we consider entire molecular networks and target critical nodes in combination.
The overarching goal of our research program is to discover and validate novel combination therapies with increased efficacy for the clinical management of HCC. Experimentally, we will take advantage of a suite of RNAi (RNA interference) tools optimized for drug target discovery and mouse models that enable in depth analysis of relevant drug targets. We have previously demonstrated that RNAi screens can be successful in discovering novel therapeutic targets for different types of cancer, including HCC. Here, we will modify the screening approach to identify genes whose inhibition cooperates with palbociclib, and establish a pipeline that can be applied to other inhibitors or other tumor types.
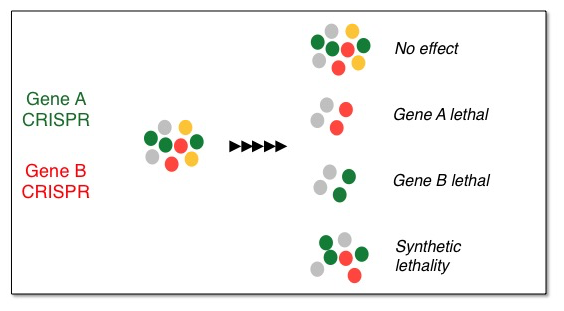
Pipeline to uncover synthetic lethal interactions in liver cancer.
Synthetic lethality arises when a combination of mutations in two or more genes leads to cell death, whereas a mutation in only one of these genes does not, and by itself is said to be viable. A synthetic lethal approach to cancer therapy is exploited to develop therapies with reduced off-target effects, and increased selectivity. We are currently developing an assay to identify synthetic lethal interactions by using the CRISPR/Cas9 system, a novel technology to genetically target genes. By linking the CRISPRs to fluorescent reporters, we have developed an assay that identifies true synthetic lethal interactions, excluding general toxicities. With these tools in hand, we aim to uncover novel synthetic interactions that can be targeted with small molecules.
Identifying novel tumor suppressor genes silenced by epigenetic mechanisms in liver cancer.
It is now recognized that cancer is both a genetic and an epigenetic disease. Genetic lesions alone cannot explain the complexity of the aberrations that arise in a cancer cell. The systematic study of genomic alterations in cancer has unveiled important cancer driver genes. However, similar studies globally studying epigenetic alterations are missing. Here, we will identify and characterize new TSGs silenced by DNA methylation in HCCs and study the role of methylated regions in cancer development. Experimentally, we will take advantage of epigenomics analyses of human and murine HCCs to inform functional studies in mice, and implement a suite of new RNAi tools and animal modeling approaches to increase the cost effectiveness and pace of our analyses. These studies will identify intrinsic mechanisms of tumor suppression that limit the progression of liver cancers, perhaps suggesting better strategies to classify these diseases and pointing towards new therapeutic targets.