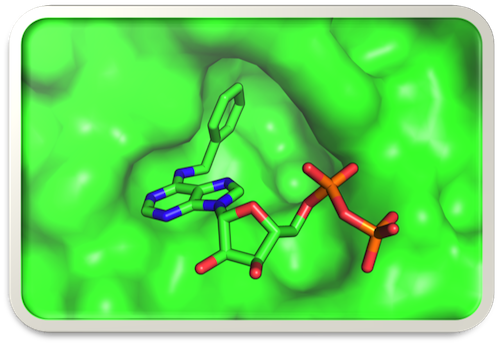
Model of the bulky analog N6-(benzyl)-ADP docked in the expanded active site of analog-sensitive (AS) mutant human Cdk2
A Network in Control of Cell Division and Gene Expression
How do cells decide when to divide and, having committed to that fate, how do they faithfully duplicate and segregate their contents—including their DNA? Among the progeny of these divisions, which all carry the same genetic information, how are different programs of gene expression established and maintained? Our lab studies the regulation of both processes—cell division and gene expression—by a network of cyclin-dependent kinases (CDKs). We take a chemical genetic approach, by expanding the ATP-binding pocket of a targeted CDK to accommodate bulky ATP analogs—both inhibitors and substrates—that cannot bind to the wild-type kinase (Figure). In multicellular organisms, cell-cycle and transcriptional CDK networks are linked by Cdk7, the CDK-activating kinase (CAK), which is also part of the general transcription factor TFIIH. We determined how Cdk7 coordinates its cell-cycle function with a second essential function during the transcription cycle of RNA polymerase (RNAP) II. Our studies reveal similarities of organization and regulatory logic between cell division and mRNA synthesis; both are cycles, in which checkpoints and surveillance systems enforce fidelity, directionality and irreversibility. By chemical genetic analysis of the CDK network we hope to define the precise roles and targets of specific CDK-cyclin complexes in cell-cycle and transcriptional regulation, and to identify potential vulnerabilities of cancer cells to specific CDK inhibitors.
