Currently, our lab will focus on three research topics:
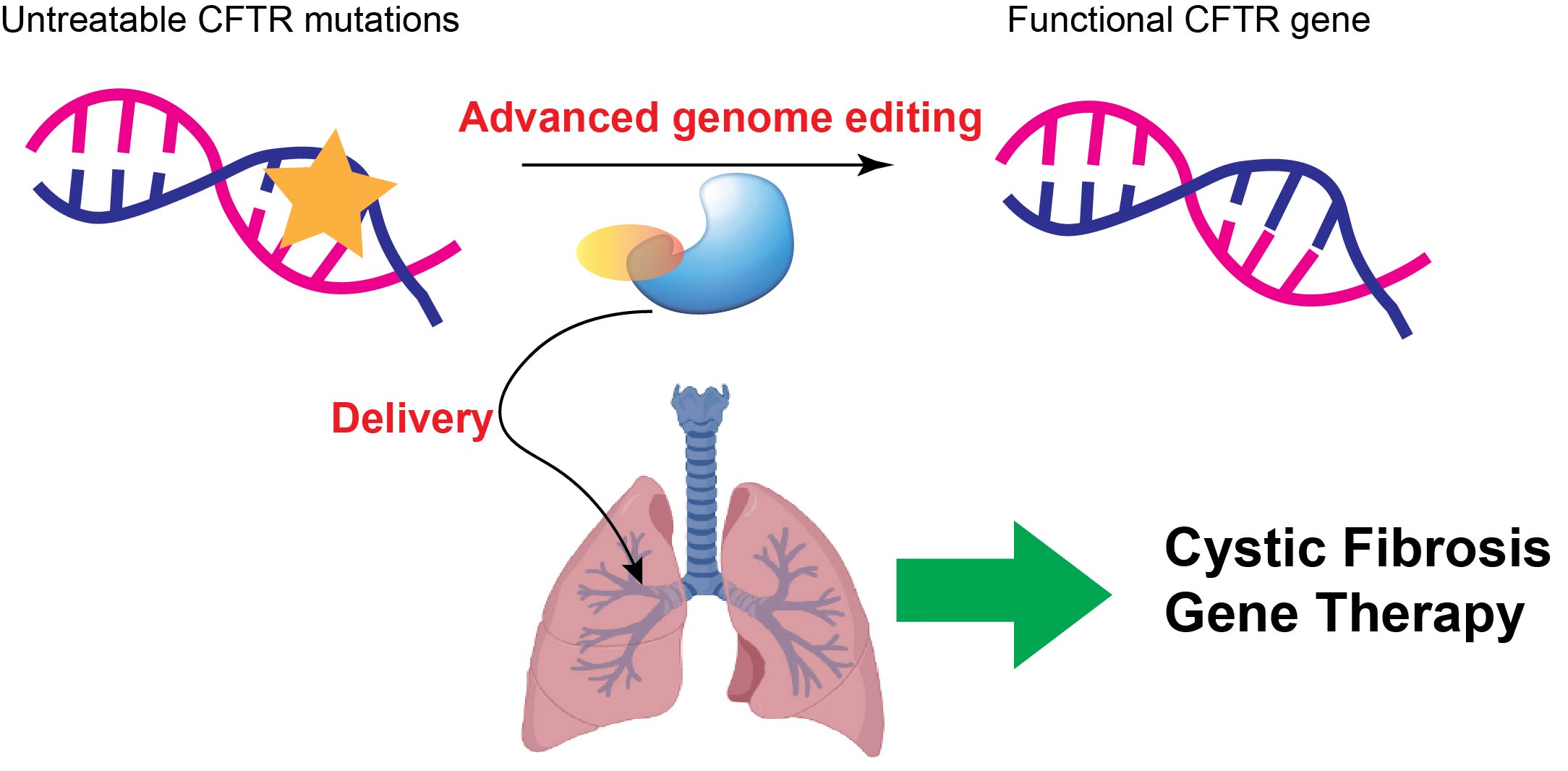
Topic 1: Developing base editing and prime editing-based gene therapies to correct untreatable cystic fibrosis mutations in the lung
Cystic fibrosis (CF) is a genetic disorder that causes damage to multiple organs. Lung infection is the primary cause of mortality in CF patients. Although four small molecule drugs have been developed and approved by FDA to treat some CF patients, around 11% of the CF patients are still untreatable because they carry unactionable mutations in the CFTR gene, the causative gene of CF. These mutations can be point mutations (creating pre-mature stop codon), insertions, and large deletions. Advanced CRISPR-based gene editors, such as base editing and prime editing, could install nearly all types of desired edits without creating a double-stranded DNA break. Thus, they hold great promise to correct those unactionable CFTR mutations. In the lab, we will engineer these tools to improve their efficiency and accuracy in editing the CFTR locus. Further, we will explore effective delivery methods to target these newly-engineered genome editors to the lung airway epithelial cells to correct lung phenotypes in the CF animal models. Our goal is to provide an effective and low-toxicity next-generation gene therapy to those untreatable CF patients.
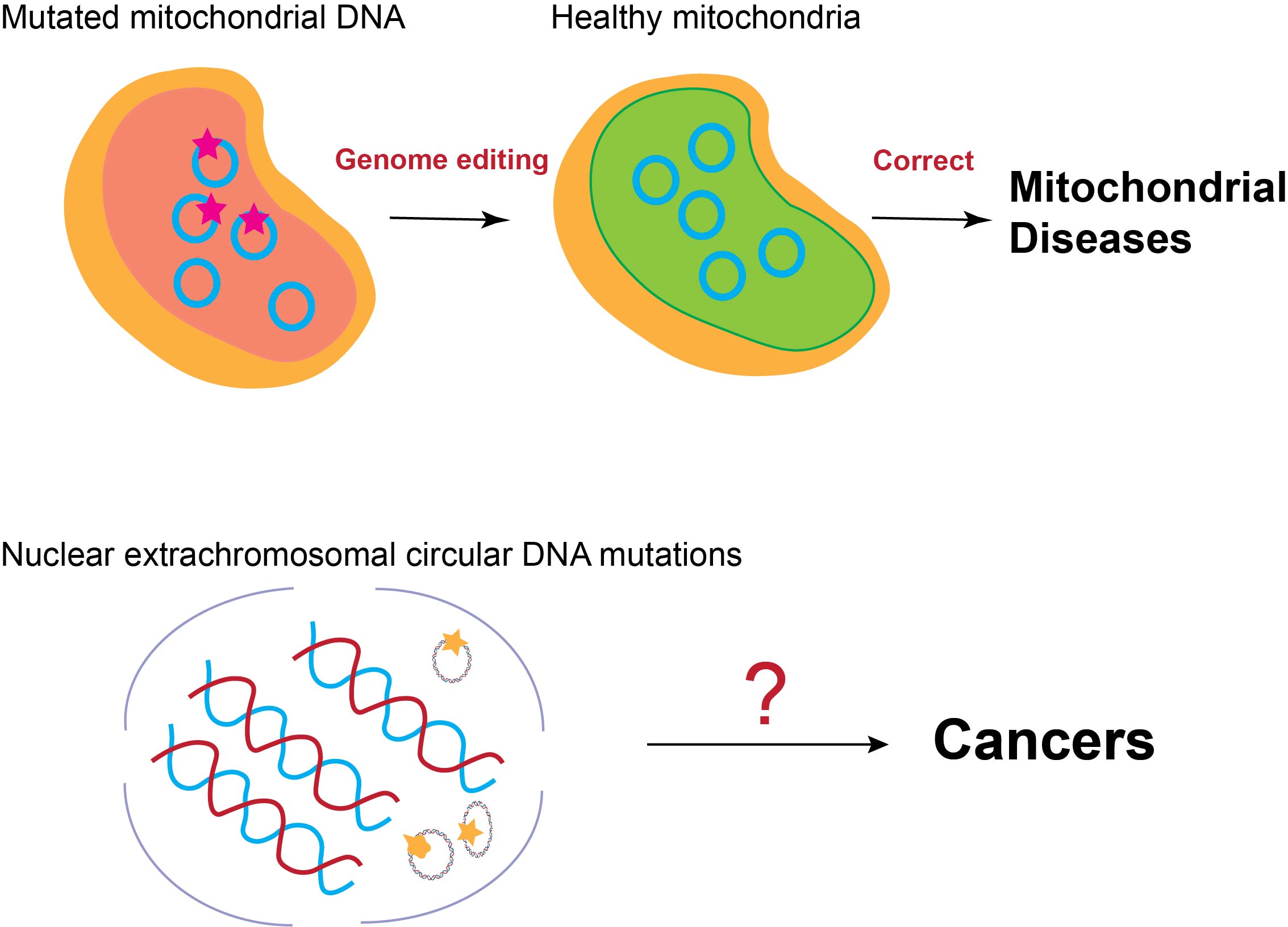
Topic 2: Investigating mutations within the Extrachromosomal DNA and relevant genetic diseases
There are different forms of circular DNA existing in mammalian cells, such as mitochondrial DNA (mtDNA) and Extrachromosomal circular DNA (eccDNA). The mutations within these circular DNA could lead to various genetic diseases. For example, mutations in eccDNA are associated with cancers, while mitochondrial DNA mutations are the major cause of mitochondrial disorders. However, due to high sequence similarity with chromosome DNA (eccDNA) and compartmentalization (mtDNA), they are hard to be manipulated, hindering the discovery of the causality between mutations and phenotype and the development of the targeted therapy. Our lab will engineer relevant CRISPR-based gene editors to directly edit these circular DNA to investigate their roles in disease pathogenesis and explore potential therapies.
Topic 3: Modulate drug resistance in liver cancers by targeting the driving mutations.
Liver cancer patients tend to develop resistance to chemotherapy and targeted therapy. These are due to the cancer genetic heterogeneity. Patient tumor samples or even cancer cell line have different subclones within one population. Some of these subclones harbor the mutations that could direct therapeutic resistance. After drug selection stress, the cancer cell subclones could repopulate the whole population. It is also possible that some subclones have plasticity, which can develop the mutations conferring resistance. To modulate these mutation-driving resistance, we will use deep sequencing to identify the enriched mutations at different time points post drug treatments. To reveal the causality between these enriched mutations and resistance, we will use genome editing method to correct these identified mutations into their wild-type counterpart to explore whether the cells will be more sensitive to the corresponding drugs. If yes, we will investigate the downstream pathways that are controlled by these mutations to confer therapeutic resistance, and further explore potential therapeutic targets to combat drug resistance in liver cancers.
