Our Research
We study cell-type-specific epigenetic regulation in the human brain in brain disorders, development, and evolution.

Stella Dracheva, PhD, Director
Professor,
Department of Psychiatry
stella.dracheva@mssm.edu
Funding and Awards
Active NIH grants:
NIMH U01MH122590 (PI: Dracheva)
2/3 High-resolution mapping of cell type-specific DNA (hydroxy)methylation in the human brain during postnatal development and in psychiatric disease
NIMH U01MH116442 (MPI: Dracheva)
The 3D genome in transcriptional regulation across the postnatal life span, with implications for schizophrenia and bipolar disorder
NIDA R01DA04324 (MPI: Dracheva)
Cell Specificity of the Human Heroin Epigenome
Active VA Merit Awards:
BX003624 (PI: Dracheva)
The Role of ADAR2-associated RNA Editing in Pathogenesis of ALS
BX002876, (PI: Dracheva)
Neuronal Subtype Specific Epigenetic Regulation in Schizophrenia
Pending VA Merit Awards:
BX005585 (PI: Dracheva)
Cell-type-specific molecular pathology of ALS in U.S. military Veterans
BX005160 (PI: Dracheva)
The role of microglia in major depressive disorder
Current Projects
Transcriptional and regulatory deficits in amyotrophic lateral sclerosis (ALS)
ALS is a devastating neurodegenerative disorder that is manifested in the degeneration of upper and lower motor neurons. In addition, in many patients the ALS pathology was reported in cortical regions outside the motor cortex (MOC), and ~10% of ALS patients also have frontotemporal dementia (FTD). To date, mutations in more than 50 genes have been linked to ALS. Hexanucleotide repeat expansion in C9orf72 (C9) is the most common cause of ALS and FTD.
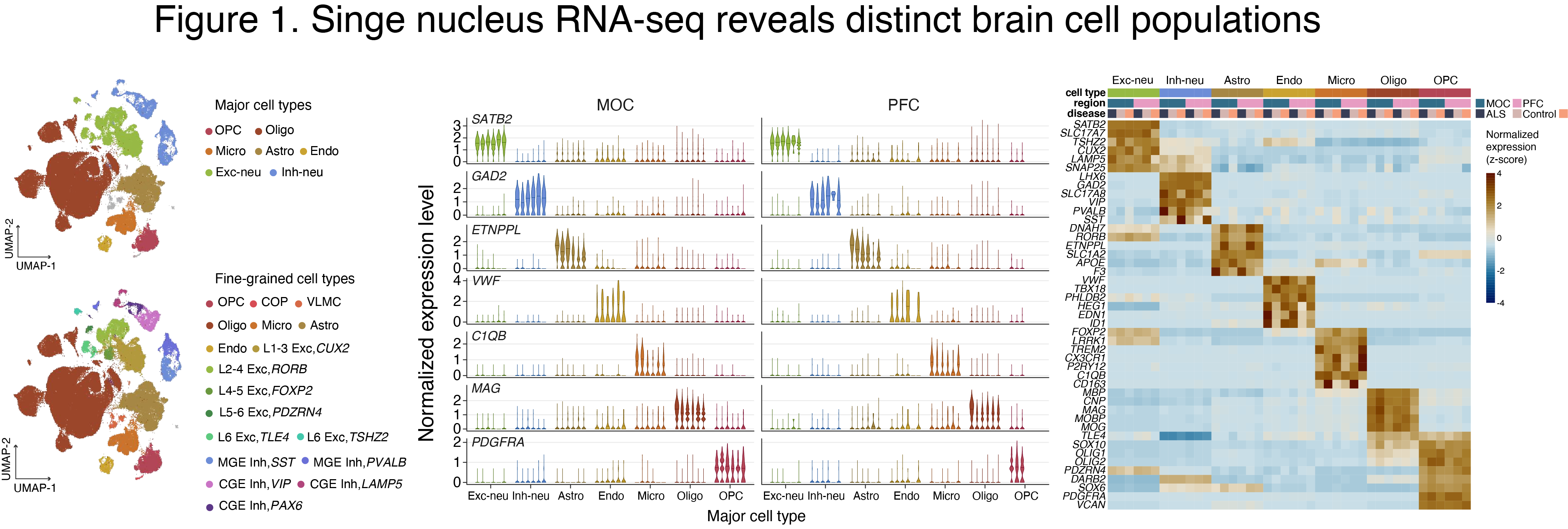
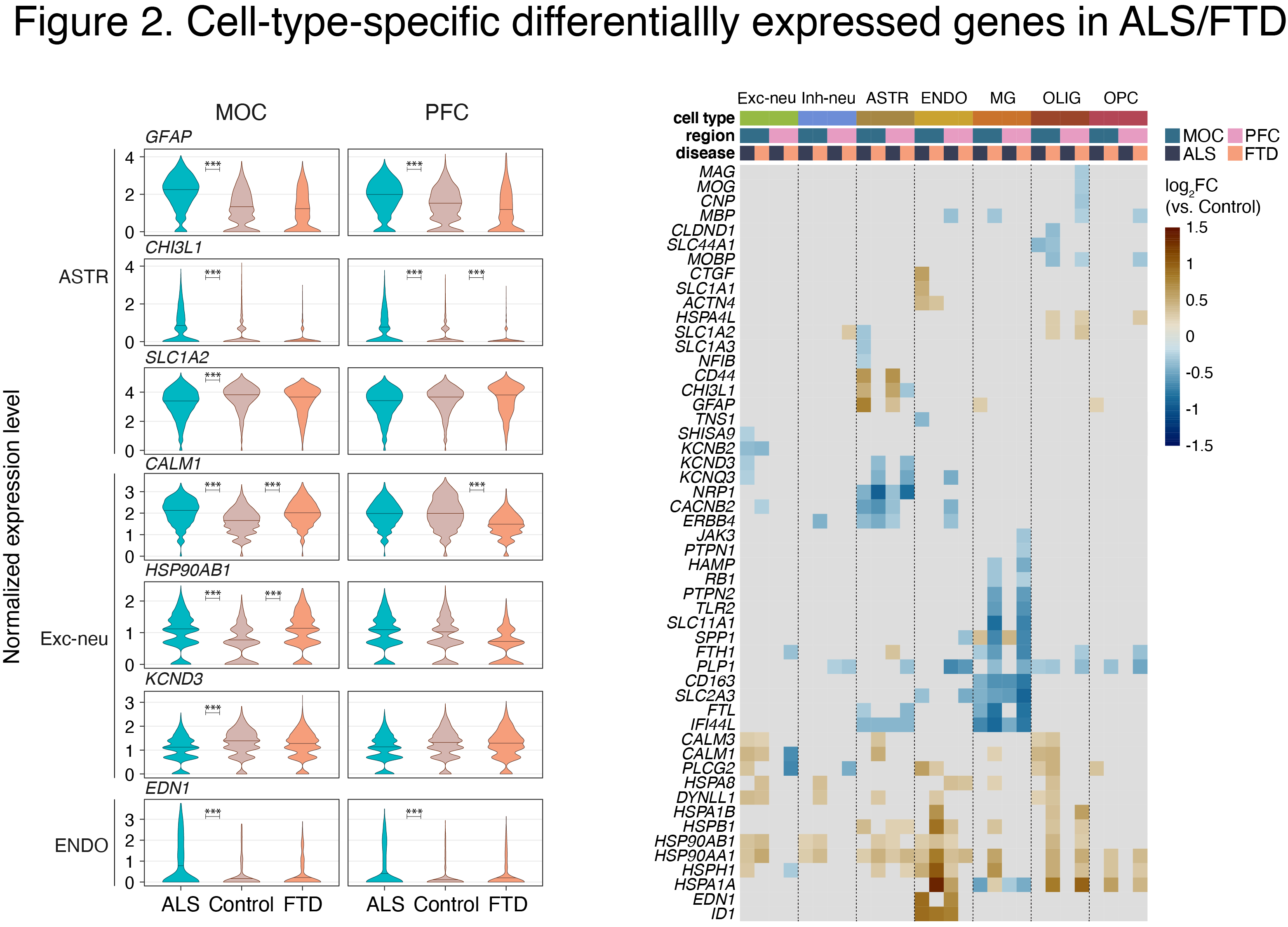
We recently performed single nucleus (sn) RNA-seq analysis using autopsied MOC and prefrontal cortex (PFC) from C9-ALS and C9-FTD patients (Fig. 1). We identified disease-related transcriptional changes in many cell types, including shared effects in ALS and FTD, and numerous disease-specific alterations (manuscript in preparation) (Fig. 2).
We are currently expanding our ALS studies:
1. We are investigating cell-type-specific regulatory landscapes in the brains of ALS patients using ChIP-seq assays of histone modifications that are indicative of active promoters and enhancers.
2. We are exploring if C9orf72 mutation perturbs a local chromatin structure, thus influencing enhancer-promoter loops in the C9orf72 locus in several different brain cell types.
3. Although ALS typically leads to death within 3 to 5 years after initial symptom onset, ~10% of patients with ALS live significantly longer (>10 years). Thus, we are elucidating cell-type-specific underpinnings of the long duration ALS.
Cell-type-specific DNA methylation (DNAm) and hydroxymethylation (DNAhm) in brain development and brain disorders
The identity of numerous brain cell types is established and maintained through spatiotemporal regulation of gene expression that, in turn, is defined by cell type-specific epigenomic marks, including DNA(h)m. Aberrant DNA(h)m has been linked to neuropsychiatric disorders, e.g., schizophrenia and bipolar disorder. In our previously published studies, we generated high-resolution maps of DNA(h)m in three major cell types in the human prefrontal cortex–glutamatergic excitatory neurons (GLU), medial ganglionic eminence-derived GABAergic inhibitory neurons (MGE-GABA), and oligodendrocytes (OLIG), together with gene expression and histone modification profiling (Kozlenkov et al. 2014, 2016, 2018). Our findings indicate differential roles for DNAm and DNAhm in regulation of gene expression in different brain cell types, with implications for etiology of human brain disorders.
In our ongoing work, we are:
(1) Exploring the dynamics of DNA(h)m and gene expression in developing and adult brain in GLU and MGE-GABA using nuclei isolated by flow cytometry-based approaches that were established in our lab (see Figure).
(2) Establishing developmental dynamics of fine neuronal subpopulations using single nucleus (sn)DNA methylomes.
Cell-type-specific regulatory evolution in human brain
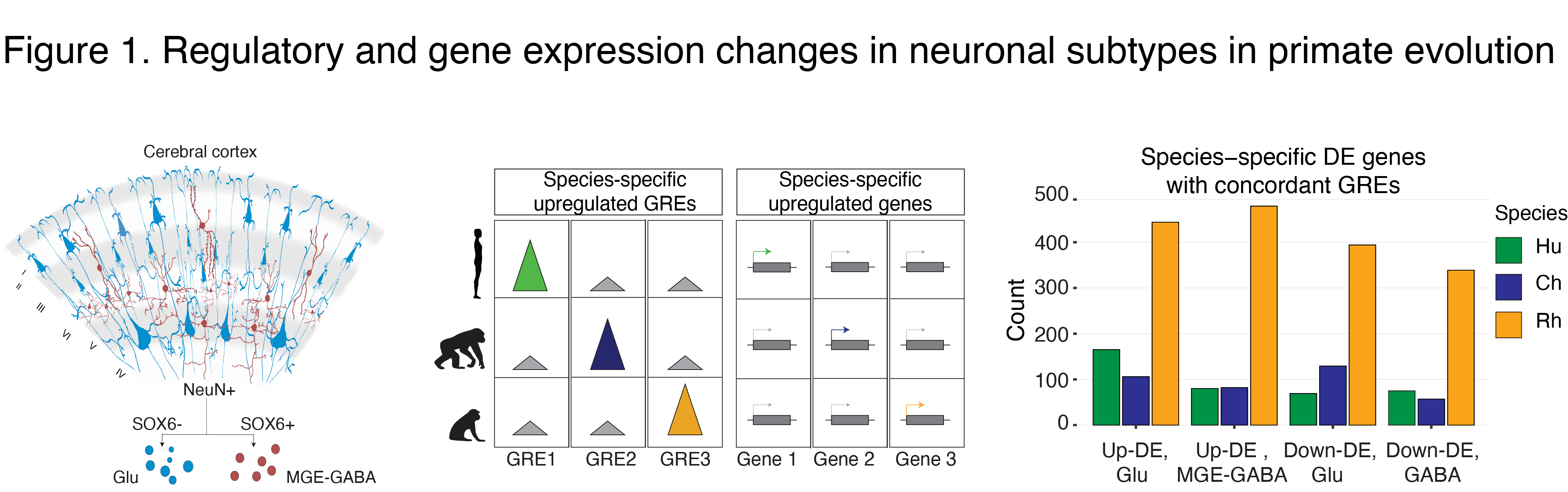
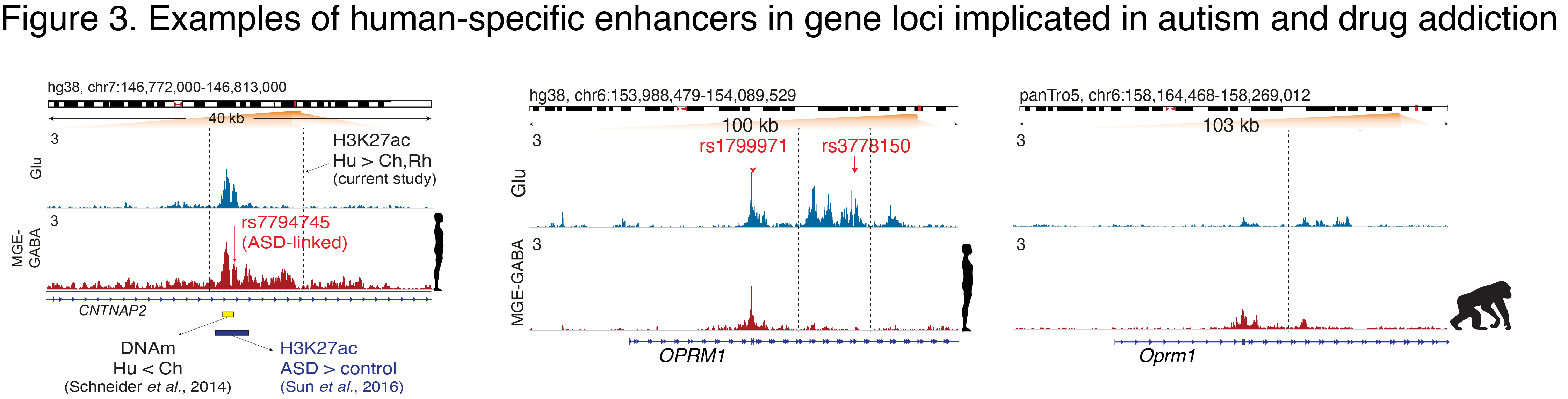
The human cerebral cortex contains many cell types that likely underwent independent functional changes during evolution. We are studying epigenomic and transcriptomic maps in different primate species in order to uncover networks that underlie the evolution of human brain function and human-specific traits. In our recent paper, we identified neuron-subtype-specific regulatory elements in human, chimpanzee and rhesus macaque prefrontal cortex that previously went undetected in bulk brain tissue (Kozlenkov et al. 2020) (Fig. 1).
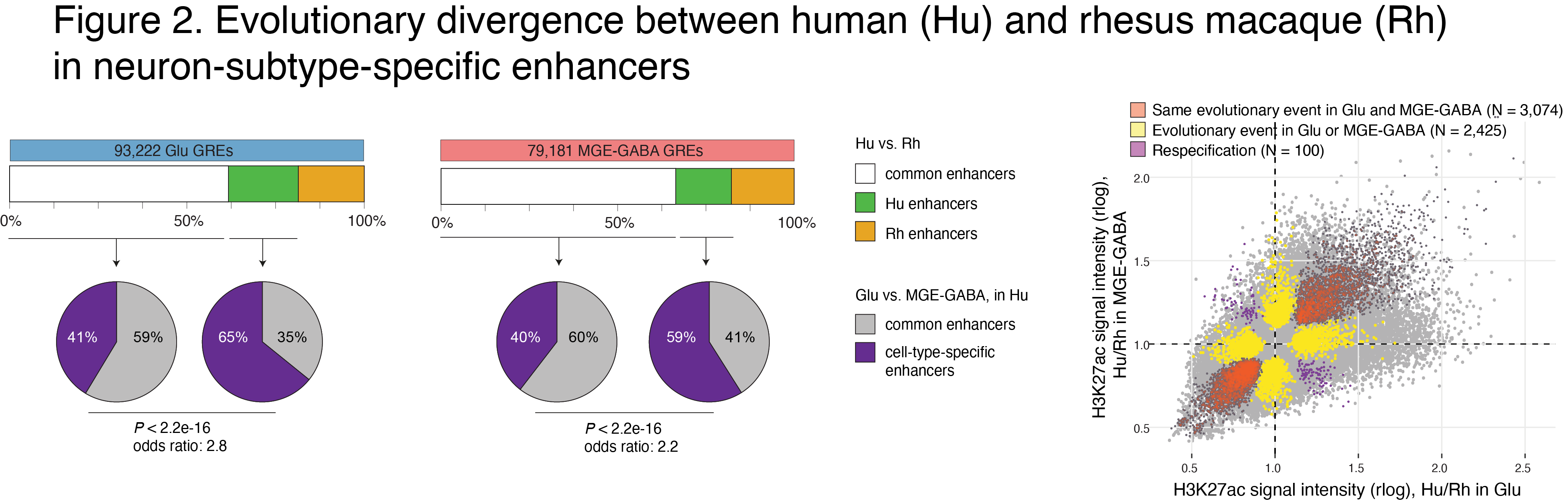
We observed preferential evolutionary divergence in neuron-subtype-specific regulatory elements and showed that a substantial fraction of pan-neuronal regulatory elements undergoes subtype-specific evolutionary changes (Fig. 2).
Human-specific regulatory changes were uncovered in multiple genes, including those associated with language, autism spectrum disorder and drug addiction (Fig. 3). This study sheds light on the interplay between regulatory evolution and cell-type-dependent gene expression programs, and provides a resource for further exploration of human brain evolution and function.
The role of microglia in major depressive disorder (MDD)
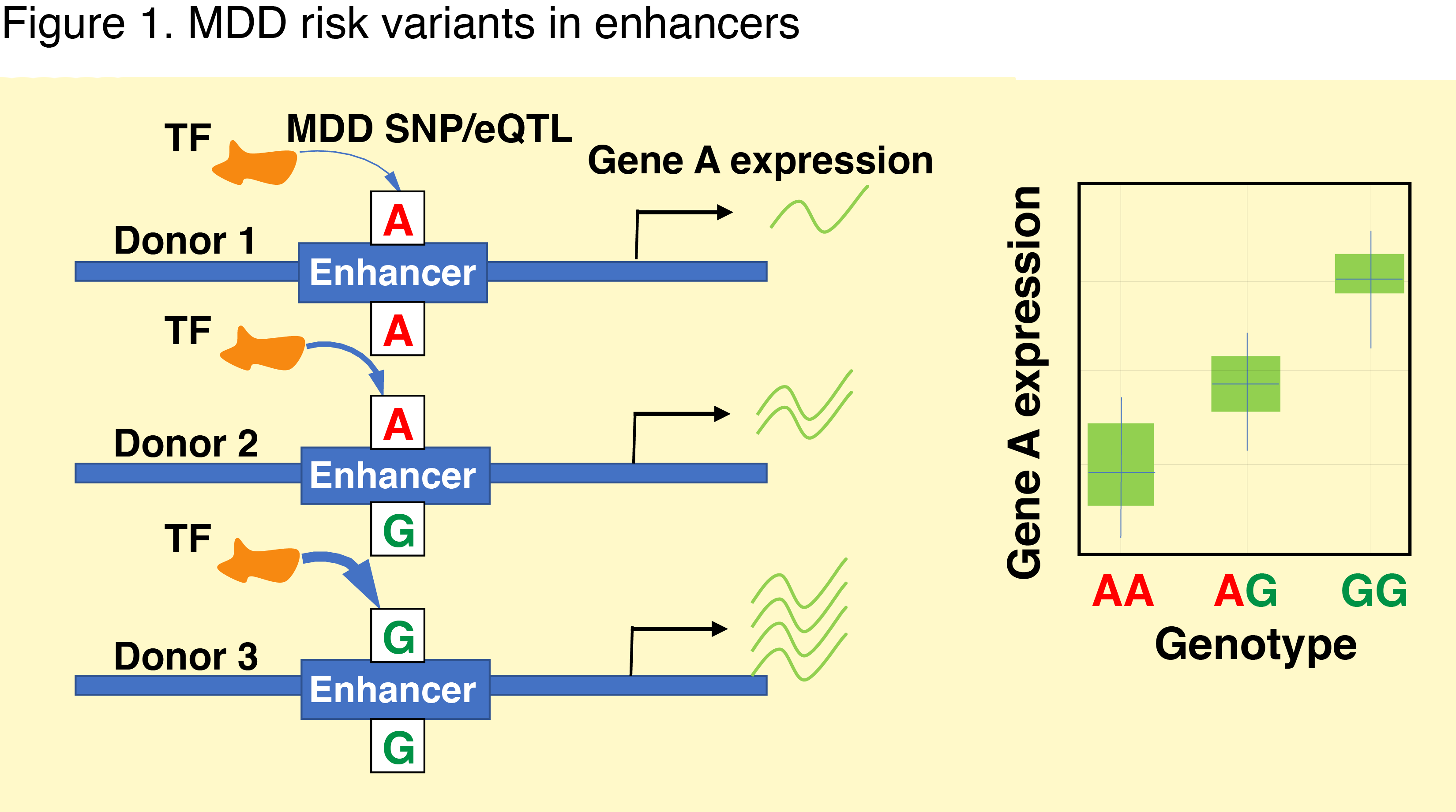
MDD affects more than 300 million people worldwide and increased risk of suicide. Available treatments are often ineffective and there is an urgent need to obtain a better understanding of the biological basis of MDD. Genome-wide association studies (GWAS) discovered > 100 MDD risk loci. The majority of them reside within non-coding regions where they are predicted to alter the activity of gene regulatory elements (GREs). These elements help to recruit transcription factors and thus to regulate the expression of nearby genes (Fig. 1). Linking the disease risk variants with expression quantitative trait loci (eQTLs) and GREs in relevant cell types yielded formidable results in other psychiatric disorders and holds promise for MDD.
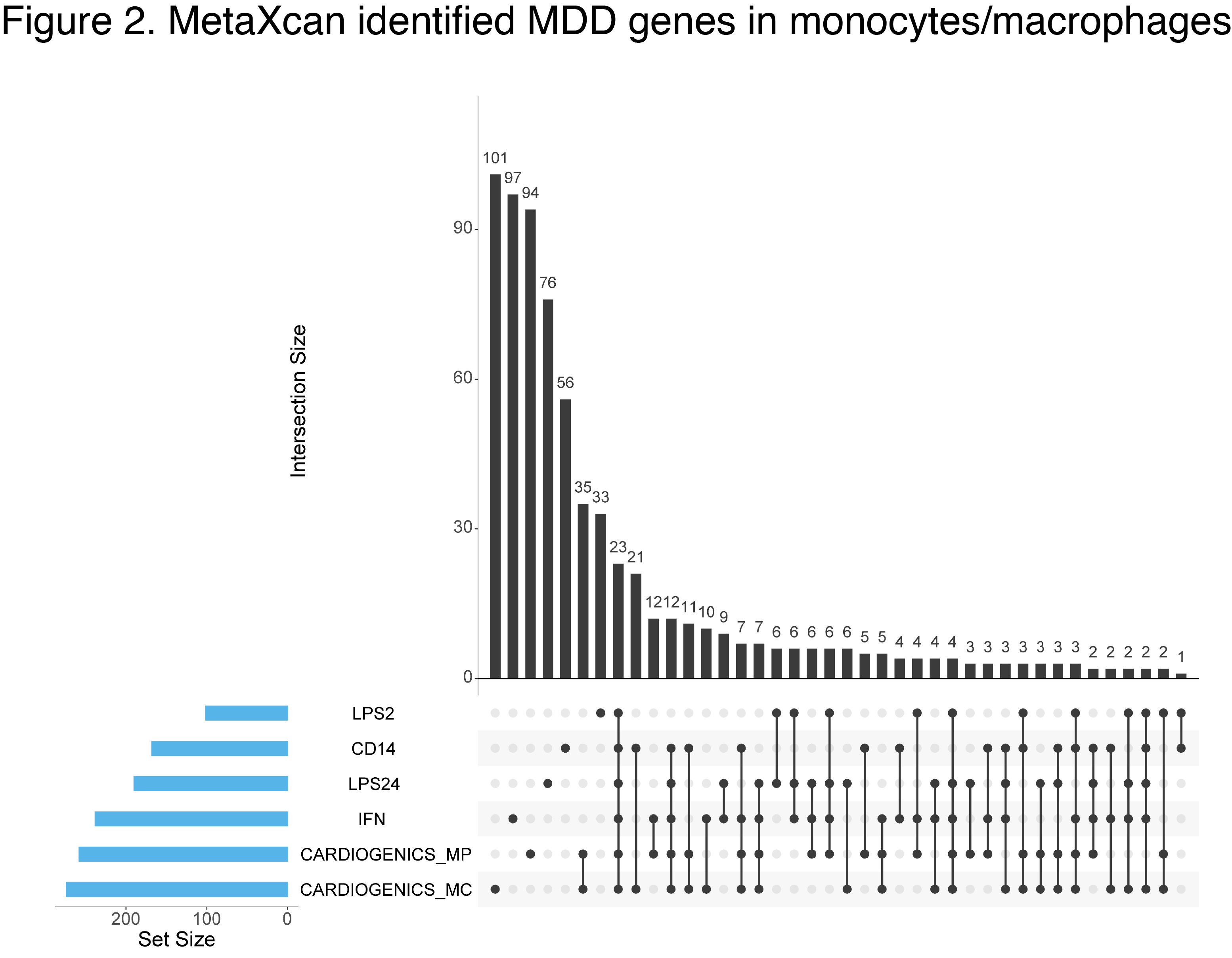
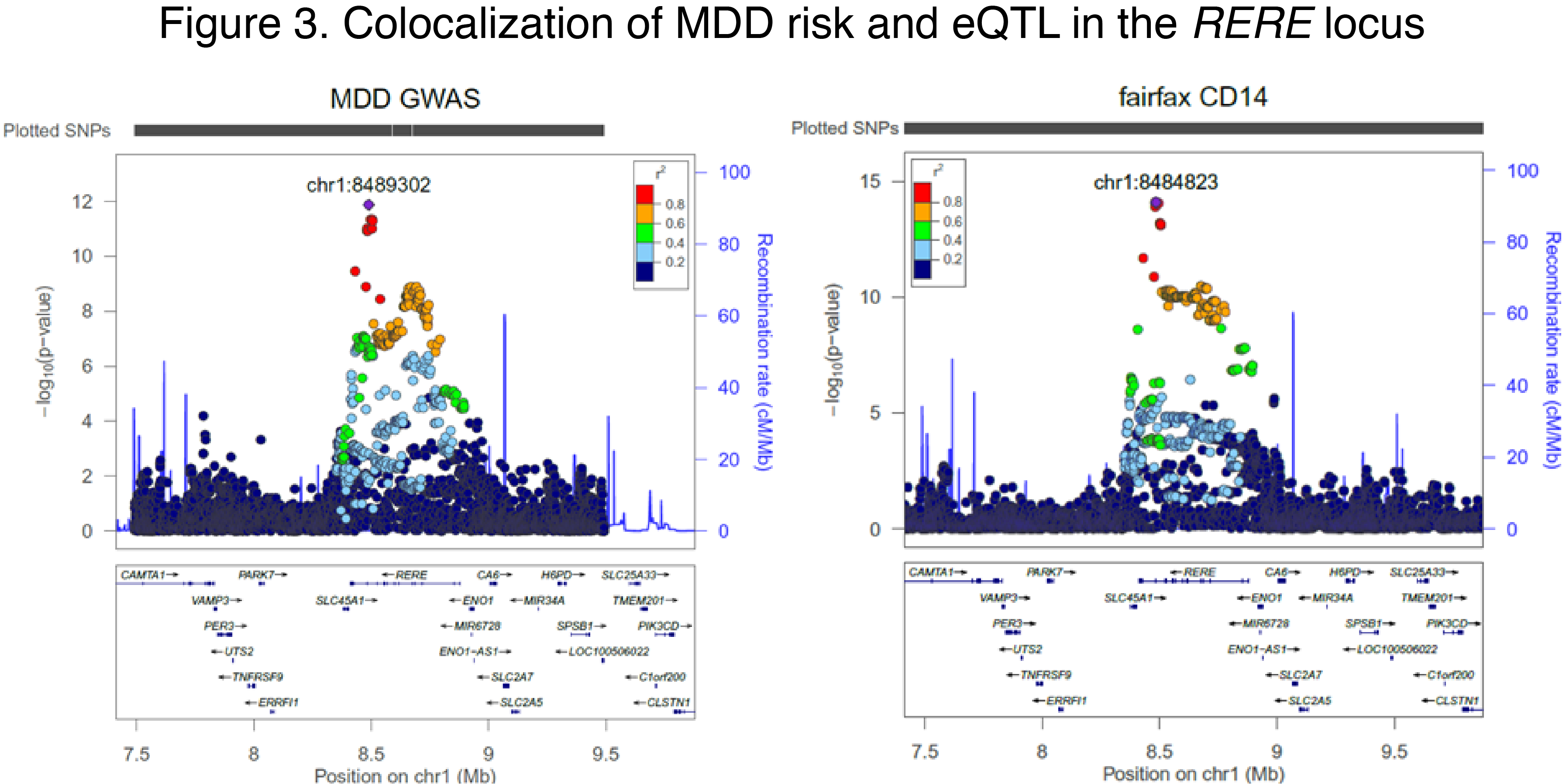
Emerging evidence implicates microglia (MG)—the immune cells of the brain—to the pathogenesis of MDD. Our preliminary data shows that MDD genetic risk variants are reflected in gene expression changes in blood-derived myeloid cells (Fig. 2-3). Because of the substantial overlap in transcriptional landscapes between these cells and MG, these data suggest that MG might play an important role in MDD. However, large-scale gene expression data in MG are not available.
Our lab has recently developed a fluorescence-activated nuclear sorting method to isolate MG nuclei from postmortem brain. We are characterizing MG transcriptome in the brains of MDD in order to map MG eQTLs and integrate them with MDD GWAS for the identification of putative causal variants and genes. The resulting regions will be validated using MG differentiated from pluripotent stem cells.
Our Team

Stella Dracheva, PhD, Director
Research Professor,
Department of Psychiatry
stella.dracheva@mssm.edu

Alexey Kozlenkov PhD
Assistant Professor, Department of Psychiatry
Alexey.Kozlenkov@mssm.edu

Manoj K. Jaiswal, PhD
Instructor, Department of Psychiatry
Manoj.Jiswal@mssm.edu

Ramu Vadukapuram, MD
Postdoctoral Fellow, Department of Psychiatry
Ramu.Vadukapuram@mssm.edu

Ping Zhou, MS
Associate Scientist, Department of Psychiatry
Ping.Zhou@mssm.edu

Shahrukh Khalique
Undergraduate trainee, Columbia University
sk4585@columbia.edu
Publications
Recent Publications
[1] Hu B, Won H, Mah W, Park R, Kassim B, Spiess K, Kozlenkov A, Crowley CA, Pochareddy S, Li Y, Dracheva S, Sestan N, Akbarian S, Geschwind DH. Neuronal and glial 3D chromatin architecture illustrates cellular etiology of brain disorders. bioRxiv. 2020:2020.05.14.096917 (accepted to Nature Communications).
[2] Kozlenkov A, Vermunt MW, Apontes P, Li J, Hao K, Sherwood CC, Hof PR, Ely JJ, Wegner M, Mukamel EA, Creyghton MP, Koonin EV, Dracheva S. Evolution of regulatory signatures in primate cortical neurons at cell-type resolution. Proc Natl Acad Sci U S A. 2020;117:28422-32.
[3] Hauberg ME, Creus-Muncunill J, Bendl J, Kozlenkov A, Zeng B, Corwin C, Chowdhury S, Kranz H, Hurd YL, Wegner M, Borglum AD, Dracheva S, Ehrlich ME, Fullard JF, Roussos P. Common schizophrenia risk variants are enriched in open chromatin regions of human glutamatergic neurons. Nat Commun. 2020;11:5581.
More Publications
[4] Factor DC, Barbeau AM, Allan KC, Hu LR, Madhavan M, Hoang AT, Hazel KEA, Hall PA, Nisraiyya S, Najm FJ, Miller TE, Nevin ZS, Karl RT, Lima BR, Song Y, Sibert AG, Dhillon GK, Volsko C, Bartels CF, Adams DJ, Dutta R, Gallagher MD, Phu W, Kozlenkov A, Dracheva S, Scacheri PC, Tesar PJ, Corradin O. Cell Type-Specific Intralocus Interactions Reveal Oligodendrocyte Mechanisms in MS. Cell. 2020;181:382-95 e21.
[5] Egervari G, Kozlenkov A, Dracheva S, Hurd YL. Molecular windows into the human brain for psychiatric disorders. Mol Psychiatry. 2019;24:653-73.
[6] McKenzie AT, Wang M, Hauberg ME, Fullard JF, Kozlenkov A, Keenan A, Hurd YL, Dracheva S, Casaccia P, Roussos P, Zhang B. Brain Cell Type Specific Gene Expression and Co-expression Network Architectures. Sci Rep. 2018;8:8868.
[7] Kozlenkov A, Li J, Apontes P, Hurd YL, Byne WM, Koonin EV, Wegner M, Mukamel EA, Dracheva S. A unique role for DNA (hydroxy)methylation in epigenetic regulation of human inhibitory neurons. Sci Adv. 2018;4:eaau6190.
[8] Gomez S, Garrido-Garcia A, Garcia-Gerique L, Lemos I, Sunol M, de Torres C, Kulis M, Perez-Jaume S, Carcaboso AM, Luu B, Kieran MW, Jabado N, Kozlenkov A, Dracheva S, Ramaswamy V, Hovestadt V, Johann P, Jones DTW, Pfister SM, Morales La Madrid A, Cruz O, Taylor MD, Martin-Subero JI, Mora J, Lavarino C. A Novel Method for Rapid Molecular Subgrouping of Medulloblastoma. Clin Cancer Res. 2018;24:1355-63.
[9] Li Y, Lucas-Osma AM, Black S, Bandet MV, Stephens MJ, Vavrek R, Sanelli L, Fenrich KK, Di Narzo AF, Dracheva S, Winship IR, Fouad K, Bennett DJ. Pericytes impair capillary blood flow and motor function after chronic spinal cord injury. Nat Med. 2017;23:733-41.
[10] Kozlenkov A, Jaffe AE, Timashpolsky A, Apontes P, Rudchenko S, Barbu M, Byne W, Hurd YL, Horvath S, Dracheva S. DNA Methylation Profiling of Human Prefrontal Cortex Neurons in Heroin Users Shows Significant Difference between Genomic Contexts of Hyper- and Hypomethylation and a Younger Epigenetic Age. Genes (Basel). 2017;8.
[11] Lu AT, Hannon E, Levine ME, Hao K, Crimmins EM, Lunnon K, Kozlenkov A, Mill J, Dracheva S, Horvath S. Genetic variants near MLST8 and DHX57 affect the epigenetic age of the cerebellum. Nat Commun. 2016;7:10561.
[12] Kozlenkov A, Wang M, Roussos P, Rudchenko S, Barbu M, Bibikova M, Klotzle B, Dwork AJ, Zhang B, Hurd YL, Koonin EV, Wegner M, Dracheva S. Substantial DNA methylation differences between two major neuronal subtypes in human brain. Nucleic Acids Res. 2016;44:2593-612.
[13] Halene TB, Kozlenkov A, Jiang Y, Mitchell AC, Javidfar B, Dincer A, Park R, Wiseman J, Croxson PL, Giannaris EL, Hof PR, Roussos P, Dracheva S, Hemby SE, Akbarian S. NeuN neuronal nuclei in non-human primate prefrontal cortex and subcortical white matter after clozapine exposure. Schizophr Res. 2016.
[14] Di Narzo AF, Kozlenkov A, Ge Y, Zhang B, Sanelli L, May Z, Li Y, Fouad K, Cardozo C, Koonin EV, Bennett DJ, Dracheva S. Decrease of mRNA Editing after Spinal Cord Injury is Caused by Down-regulation of ADAR2 that is Triggered by Inflammatory Response. Sci Rep. 2015;5:12615.
[15] Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, Geschwind DH, Mill J, Nairn AC, Abyzov A, Pochareddy S, Prabhakar S, Weissman S, Sullivan PF, State MW, Weng Z, Peters MA, White KP, Gerstein MB, Amiri A, Armoskus C, Ashley-Koch AE, Bae T, Beckel-Mitchener A, Berman BP, Coetzee GA, Coppola G, Francoeur N, Fromer M, Gao R, Grennan K, Herstein J, Kavanagh DH, Ivanov NA, Jiang Y, Kitchen RR, Kozlenkov A, Kundakovic M, Li M, Li Z, Liu S, Mangravite LM, Mattei E, Markenscoff-Papadimitriou E, Navarro FC, North N, Omberg L, Panchision D, Parikshak N, Poschmann J, Price AJ, Purcaro M, Reddy TE, Roussos P, Schreiner S, Scuderi S, Sebra R, Shibata M, Shieh AW, Skarica M, Sun W, Swarup V, Thomas A, Tsuji J, van Bakel H, Wang D, Wang Y, Wang K, Werling DM, Willsey AJ, Witt H, Won H, Wong CC, Wray GA, Wu EY, Xu X, Yao L, Senthil G, Lehner T, Sklar P, Sestan N. The PsychENCODE project. Nat Neurosci. 2015;18:1707-12.
[16] Kozlenkov A, Roussos P, Timashpolsky A, Barbu M, Rudchenko S, Bibikova M, Klotzle B, Byne W, Lyddon R, Di Narzo AF, Hurd YL, Koonin EV, Dracheva S. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Research. 2014;42:109-27.
[17] Di Narzo AF, Kozlenkov A, Roussos P, Hao K, Hurd Y, Lewis DA, Sibille E, Siever LJ, Koonin E, Dracheva S. A unique gene expression signature associated with serotonin 2C receptor RNA editing in the prefrontal cortex and altered in suicide. HumMolGenet. 2014.
[18] Lyddon R, Dwork AJ, Keddache M, Siever LJ, Dracheva S. Serotonin 2c receptor RNA editing in major depression and suicide. World J BiolPsychiatry. 2013;14:590-601.
[19] Navarrett S, Collier L, Cardozo C, Dracheva S. Alterations of serotonin 2C and 2A receptors in response to T10 spinal cord transection in rats. Neurosci Lett. 2012;506:74-8.
[20] Lyddon R, Navarrett S, Dracheva S. Ionotropic glutamate receptor mRNA editing in the prefrontal cortex: no alterations in schizophrenia or bipolar disorder. J Psychiatry Neurosci. 2012;37:267-72.
[21] Carmel L, Koonin EV, Dracheva S. Dependencies among editing sites in serotonin 2C receptor mRNA. PLoSComputBiol. 2012;8:e1002663.
[22] Lyddon R, Cuppen E, Haroutunian V, Siever LJ, Dracheva S. No link of serotonin 2C receptor editing to serotonin transporter genotype. Neuroreport. 2010;21:1080-4.
[23] Kerns D, Vong GS, Barley K, Dracheva S, Katsel P, Casaccia P, Haroutunian V, Byne W. Gene expression abnormalities and oligodendrocyte deficits in the internal capsule in schizophrenia. SchizophrRes. 2010;120:150-8.
[24] De GR, Sosa MA, Dracheva S, Elder GA. Presenilin-1 regulates induction of hypoxia inducible factor-1alpha: altered activation by a mutation associated with familial Alzheimer’s disease. MolNeurodegener. 2010;5:38.
[25] Wong K, Lyddon R, Dracheva S. TaqMan-based, real-time quantitative polymerase chain reaction method for RNA editing analysis. AnalBiochem. 2009;390:173-80.
[26] Lamprecht R, Dracheva S, Assoun S, LeDoux JE. Fear conditioning induces distinct patterns of gene expression in lateral amygdala. Genes Brain Behav. 2009;8:735-43.
[27] Dracheva S, Lyddon R, Barley K, Marcus SM, Hurd YL, Byne WM. Editing of serotonin 2C receptor mRNA in the prefrontal cortex characterizes high-novelty locomotor response behavioral trait. Neuropsychopharmacology. 2009;34:2237-51.
[28] Barley K, Dracheva S, Byne W. Subcortical oligodendrocyte- and astrocyte-associated gene expression in subjects with schizophrenia, major depression and bipolar disorder. SchizophrRes. 2009;112:54-64.
[29] Dracheva S, Patel N, Woo DA, Marcus SM, Siever LJ, Haroutunian V. Increased serotonin 2C receptor mRNA editing: a possible risk factor for suicide. MolPsychiatry. 2008;13:1001-10.
[30] Dracheva S, Chin B, Haroutunian V. Altered serotonin 2C receptor RNA splicing in suicide: association with editing. Neuroreport. 2008;19:379-82.
[31] Dracheva S, Byne W, Chin B, Haroutunian V. Ionotropic glutamate receptor mRNA expression in the human thalamus: absence of change in schizophrenia. Brain Res. 2008;1214:23-34.
[32] Byne W, Dracheva S, Chin B, Schmeidler JM, Davis KL, Haroutunian V. Schizophrenia and sex associated differences in the expression of neuronal and oligodendrocyte-specific genes in individual thalamic nuclei. SchizophrRes. 2008;98:118-28.
[33] Haroutunian V, Katsel P, Dracheva S, Stewart DG, Davis KL. Variations in oligodendrocyte-related gene expression across multiple cortical regions: implications for the pathophysiology of schizophrenia. IntJ Neuropsychopharmacol. 2007;10:565-73.