Regulation of Gene Expression During Erythropoiesis
Delineating the molecular events that confer the ability to express lineage-specific genes upon an initially uncommitted, pluripotent hematopoietic stem cell remain major questions in cell differentiation. Use of an immortalized erythroid cell line as a means to isolate genes that may be important for erythroid function allowed us to identify a novel, erythroid-specific gene, which was named EKLF (erythroid Krüppel-like factor).
Biochemical, molecular, cellular, developmental, and genetic studies in mice and humans have established that EKLF is an essential component required for globin switching and completion of the definitive erythroid program. Disorders of hemoglobin expression can lead to a variety of hemoglobinopathies, including sickle cell anemia and ß–thalassemia (Cooley’s anemia). As a result, our examination of EKLF’s mechanism of action has illuminated how it regulates the globin locus, and has provided us with a way to reconstruct EKLF so that it can potentially rectify one type of hemoglobin disorder.
Our discovery of EKLF has stimulated other investigators around the world to search for analogous genes that can work in a similar fashion to regulate unique targets in other tissues. EKLF is now the founding member (KLF1) of a family of eighteen proteins, some of which have been directly implicated in suppression of a specific subset of cancers.
Current Projects
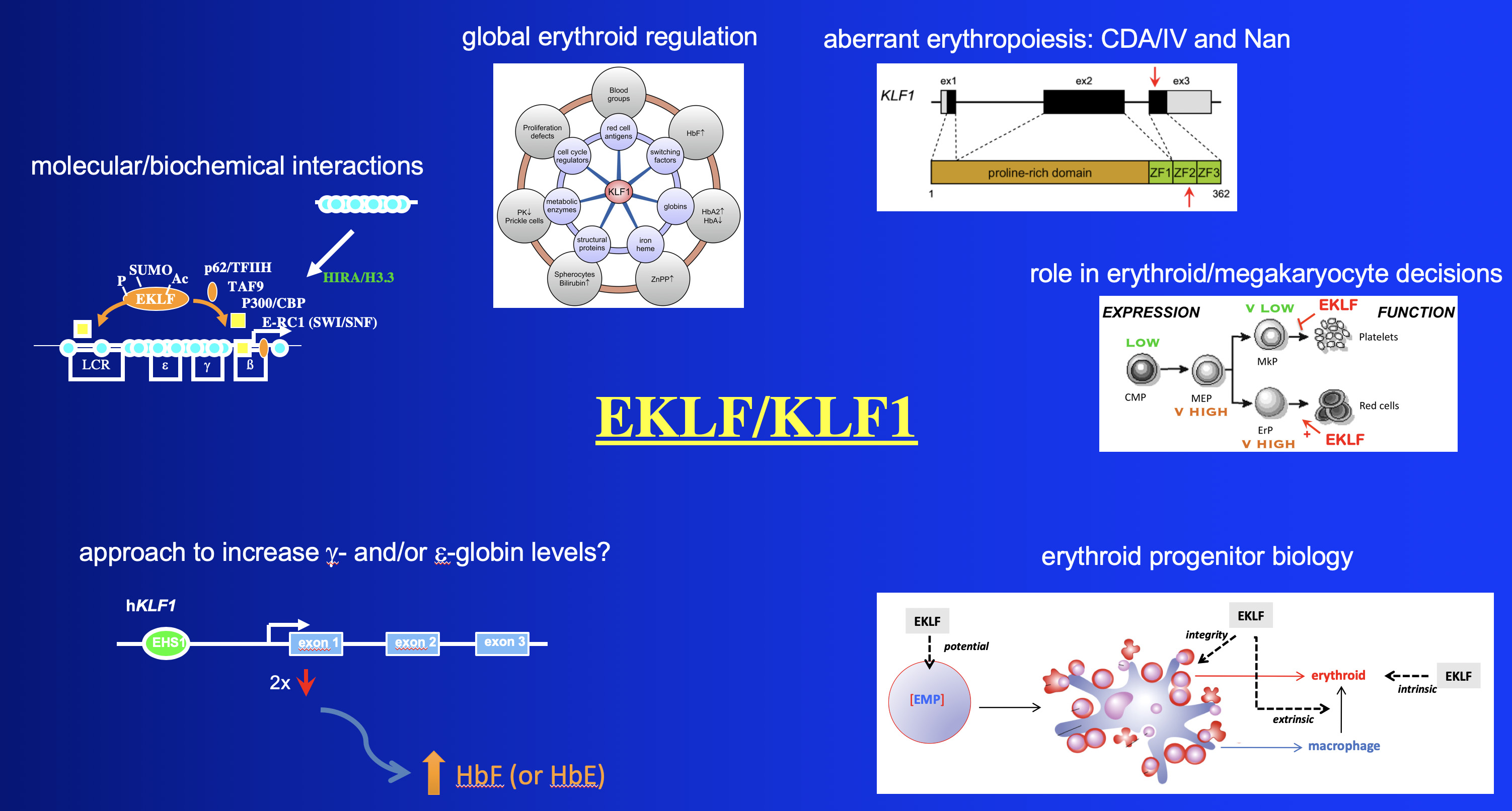
We are vigorously continuing the study of EKLF (KLF1) using a number of approaches, including biochemical and structure/function analyses of the EKLF protein, identification of its protein partners, examining its ability to extrinsically control erythropoiesis, and monitoring how EKLF expression itself is so precisely regulated during development.
Our most recent studies are focused on a number of areas:
one, a continuing analysis of EKLF protein/protein interactions and how they result in altered transcriptional and epigenetic changes at target loci;
two, on how these controls converge to regulate late events in erythropoiesis, particularly enucleation;
three, on analysis of EKLF upstream regulators to help explain its exquisite tissue-restricted expression pattern, and to possibly link alteration of its expression level to aberrant red cell biology;
four, on functional and phenotypic analyses of a neonatal anemia mouse mutant (Nan) that contains a mutation in one allele of EKLF;
finally, on determining the mechanism by which a human mutation in EKLF leads to congenital dyserythropoietic anemia.
Selected Recent Publications
Gnanapragasam MN, Jiang P, Dhara AR, Patel PN, Ramamoorthy M, Nowak RB, Fowler VM, Bieker JJ. KLF1 coordinates specialized transcriptional networks required to maintain the integrity of terminal erythropoiesis. J Cell Sci. 2025 138, jcs.264036. [highlighted in Hematopoiesis News]
Xue L*, Mukherjee K*, Kelley KA, Bieker JJ. Generation, characterization, and use of EKLF(Klf1)/CRE knock-in mice for cell-restricted analyses. Frontiers in Hematology. 2024 2, 1292589. [*co-first authors] doi: https://doi.org/10.3389/frhem.2023.1292589
Chen X*, Pillay S*, Lohmann F, Bieker JJ. Association of DDX5/p68 protein with the upstream erythroid enhancer element (EHS1) of the gene encoding the KLF1 transcription factor. J Biol Chem. 2023 299, 105489. [*co-first authors]
Identification of a genomic DNA sequence that quantitatively modulates KLF1 transcription factor expression in differentiating human hematopoietic cells. Sci Rep. 2023 13, 7589. [*co-first authors] [highlighted in Hematopoiesis News]
Mukherjee K, Bieker JJ. EKLF/Klf1 regulates erythroid transcription by its pioneering activity and selective control of RNA Pol II pause-release. Cell Rep. 2022 41, 111830.
EKLF/KLF1 expression defines a unique macrophage subset during mouse erythropoiesis. Elife. 2021 10, e61070. [highlighted in Hematopoiesis News]
A Krüppel-like factor 1 (KLF1) Mutation Associated with Severe Congenital Dyserythropoietic Anemia Alters Its DNA-Binding Specificity. Mol Cell Biol. 2020 40, e00444-19. [highlighted as an Article of Significant Interest]
Genetic disarray follows mutant KLF1-E325K expression in a congenital dyserythropoietic anemia patient. Haematologica. 2019 104, 2372-2380. [highlighted in Hematopoiesis News]
Survey and evaluation of mutations in the human KLF1 transcription unit. Sci Rep. 2018 8, 6587.
Neomorphic effects of the neonatal anemia (Nan-Eklf) mutation contribute to deficits throughout development. Development. 2017 144, 430-440. [highlighted as an In this Issue preview; highlighted in Hematopoiesis News]
EKLF/KLF1-regulated cell cycle exit is essential for erythroblast enucleation. Blood. 2016 128, 1631-41. [highlighted in Hematopoiesis News]
The DEK Oncoprotein Is a Critical Component of the EKLF/KLF1 Enhancer in Erythroid Cells. Mol Cell Biol. 2015 35, 3726-38. [*co-first authors] [highlighted in Exp Hem 43, 827 (15)]
Transcription factor EKLF (KLF1) recruitment of the histone chaperone HIRA is essential for β-globin gene expression. Proc Natl Acad Sci U S A. 2014 111, 13337-42. [highlighted in Hematopoiesis News]
Extrinsic and intrinsic control by EKLF (KLF1) within a specialized erythroid niche. Development. 2014 141, 2245-54.
Selected Reviews
Severe anemia caused by dominant mutations in Krüppel-like factor 1 (KLF1). Mutat Res. 2020 786, 108336. [highlighted in Hematopoiesis News]
Orchestration of late events in erythropoiesis by KLF1/EKLF. Curr Opin Hematol. 2017 24, 183-190.
Krüppeling erythropoiesis: an unexpected broad spectrum of human red blood cell disorders due to KLF1 variants. Blood. 2016 127, 1856-62.
KLF1: when less is more. Blood. 2014 124, 672-3.
EKLF/KLF1, a tissue-restricted integrator of transcriptional control, chromatin remodeling, and lineage determination. Mol Cell Biol. 2013 33, 4-13.
The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood. 2011 118, 2044-54.
Putting a finger on the switch. Nat Genet. 2010 42, 733-4.
The erythroblastic island. Curr Top Dev Biol. 2008 82, 23-53.
Krüppel-like factors: three fingers in many pies. J Biol Chem. 2001 276, 34355-8.
Team

Kaustav Mukherjee
Assistant Professor
kaustav.mukherjee@mssm.edu

Sanjana Pillay
Postdoctoral Fellow
sanjana.pillay@mssm.edu

Tasleem Arif
Senior Scientist
tasleem.arif@mssm.edu

Li Xue
Associate Scientist
li.xue@mssm.edu

Antanas Planutis
Lab Coordinator
antanas.planutis@mssm.edu
We use biochemical, molecular, cellular, and developmental approaches to illuminate transcriptional and epigenetic controls that lead to regulated erythroid gene expression. Our focus is on both progenitors and late red cell stages, and in determining whether select transcription factor mutations are causative for aberrant or malignant mammalian hematology.
PI Collaborators

Anna Rita Migliaccio, PhD
Professor of Histology and Embryology
Università Campus Bio-Medico di Roma
Italy
a.migliaccio@unicampus.it

Jeffrey Glassberg, MD, MA
Professor
Emergency Medicine, Hematology and Medical Oncology
Mount Sinai School of Medicine
jeffrey.glassberg@mountsinai.org

James Palis, MD
Professor
Pediatric Hematology/Oncology
University of Rochester Medical Center
Rochester, NY 14642
James_Palis@URMC.Rochester.edu

Jan Frayne, PhD
Professor
Molecular Cell Biology
University of Bristol
United Kingdom
Jan.Frayne@bristol.ac.uk

Eric E Bouhassira, PhD
Professor
Cell Biology and Medicine
Albert Einstein College of Medicine
Bronx, NY 10461
eric.bouhassira@einsteinmed.org
Alumni
Aimola, Idowu
COORDINATOR
idowuaimola@gmail.com
Africa Center Of Excellence On Neglected Tropical Disease
Ahmadu Bello University
Department of Biochemistry
Zaria Nigeria 810000
Chen, Joy
ASSOCIATE DIRECTOR
REGULATORY AFFAIRS
joy.chen@modernatx.com
Moderna
Cambridge, MA
Chen, Xiaoyong
ASSOCIATE RESEARCH SCIENTIST
xiaoyong.chen@yale.edu
Yale University School of Medicine
Dept of Pediatrics
333 Cedar St
New Haven, CT 06520
Chinta, Sree
MEDICAL STUDENT
src194@njms.rutgers.edu
Rutgers New Jersey Medical School
185 South Orange Ave
Newark NJ 07103
Dangeti, Mohan
R&D SYSTEMS ENGINEER
mohan.nimai@gmail.com
Ascensia Diabetes Care
Valhalla, NY 10595
Gruzglin, Eugenia
VP STRATEGY
gena.gruz@gmail.com
UNIFIED, an Omnicom Company
New York, NY
Gnanapragasam, M Nithya
ASSISTANT PROFESSOR
m.gnanapragasam@csuohio.edu
Cleveland State University
Department of Biological, Geological, and Environmental Sciences
Cleveland, Ohio 44115
Lohmann, Felix
RESEARCH INVESTIGATOR I
felix.lohmann@novartis.com
Novartis Institutes for BioMedical Research
Basel
Switzerland
Lopingco, Tina
VP, MEDICAL STRATEGY DIRECTOR
Tina.Lopingco@prohealthmeded.com
Neon
1400 Broadway, Floor 3
New York, NY 10018
Manwani, Deepa
PROFESSOR OF PEDIATRICS
Director of Hematology
dmanwani@montefiore.org
Albert Einstein College of Medicine
Children’s Hospital at Montefiore
Pediatric Hematology/Oncology
3415 Bainbridge Ave
Bronx, NY 10467-2490
Miller, Ira J
ASSOCIATE PROFESSOR
Ira_Miller@rush.edu
Rush University Medical Center
Dept of Pathology
1653 W Congress Pkwy
Chicago, IL 60612
Quadrini, Karen
ASSOCIATE DIRECTOR
BIOMARKERS AND PRECISION MEDICINE
KQuadrini@passagebio.com
Passage Bio
One Commerce Square
2005 Market Street, 39th Floor
Philadelphia, PA 19103
Siatecka, Mirka
ASSOCIATE PROFESSOR
msiatecka@amu.edu.pl
Adam Mickiewicz University
Institute of Experimental Biology
Department of Genetics
Umultowska 89
61-614 Poznań
Poland
Sengupta, Tanushri
RESEARCH ASSOCIATE
t-sengupta@northwestern.edu
Northwestern University
International Institute for Nanotechnology
2145 Sheridan Rd
Evanston, IL 60208
Soni, Shefali
ASSOCIATE PROGRAM OFFICER
ssoni@helmsleytrust.org
Helmsley Charitable Trust
IBD/Crohn’s Disease Program
230 Park Avenue
New York, NY 10169
Stranahan, Alec
ASSOCIATE, BIOTECH EQUITY RESEARCH
als2064@med.cornell.edu
Bank of America Merrill Lynch
New York, NY 10036
Yien, Yvette
ASSOCIATE PROFESSOR
yieny@pitt.edu
University of Pittsburgh
Department of Medicine
Vascular Medicine Institute
Pittsburgh, PA 15213
Zhang, Wenjun
SENIOR RESEARCH INVESTIGATOR II
Wenjun.Zhang@bms.com
Bristol Myers Squibb
311 Pennington Rocky Hill Rd
Pennington, NJ, 08534
Related links
Globin gene server
http://globin.cse.psu.edu/globin/
American Society of Hematology
https://www.hematology.org/
Cooley’s Anemia Foundation
https://www.thalassemia.org/
Statistics
http://www.physics.csbsju.edu/stats/
The KLF community
https://www.linkedin.com/groups/8108442/
ErythronDB
https://www.cbil.upenn.edu/ErythronDB/
Gene annotation portal
http://biogps.org/#goto=welcome
Contact Us
Location
Annenberg 2584
Phone: 212-241-5067
Office: 212-241-5067
Lab: 212-241-4143
James.bieker@mssm.edu
Mailing Address
Mount Sinai School of Medicine
Box 1020
One Gustave L Levy Place
New York, NY 10029
Postdoctoral positions are currently available. Please email a CV, a summary of past experience, and a brief statement of research interests along with the names of two to three references.
Mailing Address
Mount Sinai School of Medicine
Box 1020
One Gustave L Levy Place
New York, NY 10029

