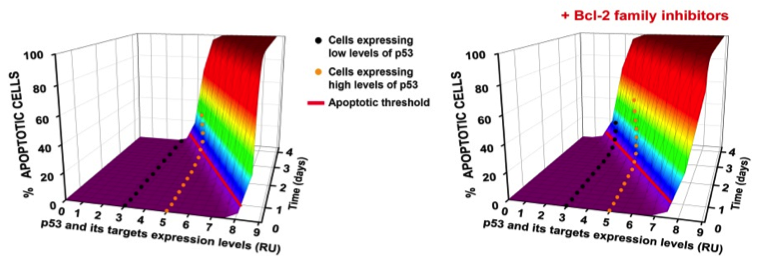
3D model for the regulation of p53 apoptotic threshold in human cells (Kracikova et al., Cell Death and Differentiation, 2013).
P53 Regulators and Effectors
The p53 tumor suppressor gene encodes a transcription factor, which is activated by DNA damage and other oncogenic stresses, influencing cell fate decisions ranging from transient growth arrest, to senescence or apoptosis by a threshold mechanism, which can be influenced by cell context as well as cellular stresses (Kracikova et al, Cell Death and Differentiation, 2013). Recent evidence indicates that p53 also contributes to a number of other homeostatic stress responses with pro-survival benefits for the organism. For example, we have shown that p53 contributes to innate immunity by enhancing IFN-dependent antiviral activity, and we are exploring other novel p53 contributions to innate immunity. How p53 regulates expression of its target genes and cell fate decisions is also an important area of investigation within our lab. In efforts to elucidate how p53 regulates its target genes, we recently identified SUV39H1, a histone code ‘writer’ responsible for the histone H3 Lys9 trimethylation (H3K9me3) repressive mark as an indirect target of down regulation by p53. As a result of p53 activation, in addition to p53 dependent loss of repressive epigenetic marks, its target promoters become enriched with the H3K4me3 epigenetic mark as well as its readers. We have shown that these epigenetic factors function in concert with P53 promoter occupancy to activate RNAPII by aiding the formation of a stable transcription pre-initiation complex. Present studies are aimed at identifying other epigenetic factors, which contribute to p53 activation of its pro-apoptotic target genes with the goal of improving therapeutic efficacy of chemotherapeutic agents for wild type p53 containing tumor cells.
